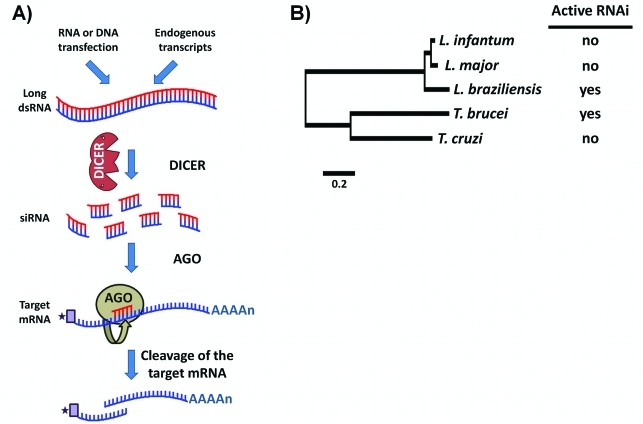
Figure 4.
Retention and loss of RNAi genes in trypanosomatids. (A) Schematic representation of the mechanism of RNA interference (RNAi) present in some trypanosomes. Double-stranded RNA (dsRNA) derived endogenously or by transfection procedures is processed by an RNase III enzyme known as DICER into small interfering dsRNA molecules (siRNA) 19–23 nt long. siRNAs associate with Argonaute (AGO), the catalytic core of the RISC (RNA-induced silencing complex), with one siRNA strand being released and the guide siRNA strand then mediating the degradation or inhibiting the translation of the target mRNA. Genes encoding components of the RNAi machinery occur only in T. brucei and in a sub-group of Leishmania. (B) Phylogenetic analysis of trypanosomatid species based on the predicted protein sequences of the housekeeping gene GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and ubiquitin. The neighbor-joining tree was generated using sequences found in the parasite genome databases (www.genedb.org) and MEGA4 software. The absence or presence of genes encoding components of active RNAi machinery (as identified by Lye et al., 2010) is indicated on the right.