Abstract
We describe a family with a history of breast and ovarian cancer in which MLPA analysis of the BRCA1 gene pointed to a deletion including a part of exon 11. Further characterization confirmed a loss of 374 bp in a region completely covered by conventional sequencing which had not revealed the deletion. Because this alteration was only detected serendipitously with an MLPA probe, we calculated the probabilities of detecting medium-sized deletions in large exons by methods including initial PCR amplification. This showed that a considerable fraction of medium-sized deletions are undetectable by currently used standard methods of mutation analyses. We conclude that long, widely overlapping amplicons should be used to minimize the risk of missing medium-sized deletions. Alternatively, large exons could be completely covered by narrow-spaced MLPA probes.
Keywords: mutation analysis, DNA sequencing, PCR, hereditary breast cancer, model calculations
Numerous different mutations in the BRCA1 and BRCA2 genes (Miki et al., 1994; Wooster et al., 1995) have been identified in families with a history of breast and ovarian cancer (King et al., 2003). Because of their broad distribution, at least all exons and adjacent sequences have to be covered by genetic testing. To identify substitutions and small insertions or deletions, PCR amplification followed by sequencing is still considered the diagnostic gold standard. In addition, MLPA (multiplex ligation-dependent probe amplification) analyses (Hogervorst et al., 2003) can reveal large deletions and rearrangements not detectable by sequencing. To reduce the time required for testing, pre-screening methods like denaturing gradient chromatography (Arnold et al., 2002) or analyses of melting curves of PCR products (van der Stoep et al., 2009) are used. These techniques, however, are still based on PCR-amplified DNA and, compared to sequencing, their sensitivities are lower. As a consequence of PCR amplification as a first step in these sorts of analyses, medium-sized heterozygous deletions affecting the binding site of one PCR primer are not recognized. Although most molecular geneticists are aware of this problem, calculations of the number of potentially missed deletions have not been published.
Here we describe a 374 bp deletion not detected in a large exon by sequencing that has been found only because the deleted region was, by chance, covered by an MLPA probe. By model calculations, we determined the likelihood to find such medium-sized deletions and suggested measures to increase it.
Clinicogenetic counselling was performed at the Familial Breast and Ovarian Cancer Center in Ulm. Genetic testing including the BRCA1 and BRCA2 genes was dependent on the risk category assigned to the family. In addition, individual risks were calculated with Cyrillic 2.1 pedigree analyses software (Cyrillic Software, Wallingford, UK).
Genomic DNA was isolated from blood with a Blood & Cell Culture DNA Maxi Kit (Qiagen, Hilden, Gemany) and all exons and adjacent regions of the BRCA1 and BRCA2 genes were amplified by standard PCR with Fast Start Taq Polymerase (Roche Diagnostics, Mannheim, Germany). End of intron 10 and beginning of exon 11 was covered by primers E11-F1 (5′-GATTTCCACCTCCAAG GTGTATGA-3′) and E11-R1 (5′-GGGAGTCCGCCTAT CATTACAT-3′). Sequencing of the purified PCR products from both directions was performed according to the suppliers’ instructions with the Dye Terminator Cycle Sequencing Kit (Beckman Coulter, Krefeld, Germany). The generated fragments were purified (Agencourt CleanSEQ, Beckman Coulter) and analysed by means of a CEQ 8000 sequencer (Beckman Coulter). Sequences were compared to the BRCA1 reference cDNA sequence (NM_007294) using Mutation Surveyor software (Softgenetics, State College, PA, USA). To describe mutations, positions are calculated from the translation initiation codon ATG (A = 1).
MLPA analysis of the BRCA1 gene was done according to the protocol supplied by the manufacturer of the SALSA-MLPA Kit P002 (MRC-Holland, Amsterdam, Netherlands) and the products were analysed with the CEQ 8000 Gene Analysis System (Beckman Coulter).
The Long-Range PCR Kit from Qiagen was used for PCR amplification with 100 ng of genomic template DNA in a total reaction volume of 50 μL under the following conditions: 3 min at 93 °C followed by 35 cycles of 15 s at 93 °C, 30 s at 58 °C and 4 min at 69 °C. Products were analysed by standard gel electrophoresis in 1% agarose gels. Forward primer E10-F1 (5′-TAAGATTGGTCAGCTTT CTGTAATC-3′, end of intron 9) was used in combination with exon 11 reverse primers E11-R1, E11-R4 (5′-TCTA TTGGGTTAGGATTTTTCTCA-3′), E11-R7 (5′-TTTTG CCTTCCCTAGAGTGCTAAC-3′), E11-R11 (5′-AGAT GCATGACTACTTCCCATAGG-3′), or E11-R13 (5′-TATTTGCAGTCAAGTCTTCCAA-3′).
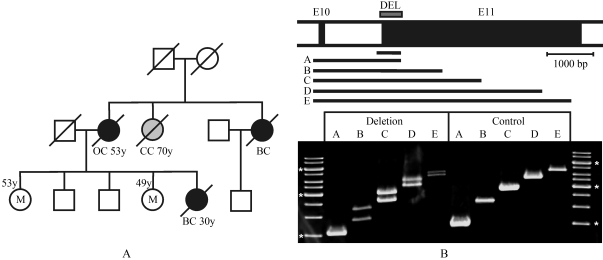
Genetic counselling and subsequent analyses of predisposing genes was requested by a 53 years old woman on the basis of her sister’s early-onset breast cancer and other cases of breast and ovarian cancer in her family (Figure 1A). Accordingly, her risk of developing breast cancer was calculated to be 31%. By sequencing of all exons and adjacent regions of the BRCA1 and BRCA2 genes, no pathogenic mutation was detected. Subsequent MLPA analyses showed a substantially reduced signal resulting from only one of two MLPA probes binding to BRCA1 exon 11, suggesting a deletion including a region close to the beginning of this exon. Long PCR with primers from the end of exon 10 to various positions of exon 11 confirmed a heterozygous medium-sized deletion (Figure 1B). These PCR products were subjected to sequence analysis which showed a deletion (del c.671-26_1018) of 374 bp. This deletion affected the binding site of one of the primers used to amplify the DNA prior to sequencing for which, as a consequence, only PCR products from the normal allele were obtained. This case raised the question as to the likeliness of detecting such medium-sized deletions by routine methodology.
Figure 1.
Detection of a medium-sized deletion in the BRCA1 gene. A: Family with a history of breast and ovarian cancer in which a deletion in BRCA1 was found. Two sisters of a woman who had developed breast cancer at age 30 harboured this mutation (M), indicated are manifestations of breast cancer (BC), ovarian cancer (OC) and colon cancer (CC). B: Characterization of the 374 bp deletion in exon 11 by long PCR. Amplified genomic regions included exon 10 (E10) and the following intron and parts of exon 11 (E11), as shown in the top scheme. The deletion in one allele resulted in double bands of longer PCR products but not of the smallest one, which was generated with the reverse primer also used in routine sequencing. The amplicon used for sequence analysis is shown in the scheme above the products of long PCR (A-E). Marker fragments labelled with an asterisk are 1500 bp, 3000 bp and 6000 bp.
Currently used strategies to detect mutations are justified by two facts: (a) Substitutions and very small insertions and deletions are most likely detected in PCR-amplified DNA by sequencing or pre-screening methods, and (b) most of the large deletions are found by MLPA. As from the case described herein we learned that a considerable fraction of medium-sized deletions might not be detected by conventional approaches, we performed model calculations to estimate the potential relevance of this problem. The simplified model exon of length l was divided into a number (q) of equally spaced PCR amplicons with a constant overlap of length a. All primers were assumed to be of the same length p and all possible deletions of length d to occur with the same likelihood over the entire region.
Short deletions, i.e. from single nucleotides to a size smaller than the region between primers in the overlap of amplicons (1 < d << a - 2p), can only lead to suppression of the mutated allele if one of the terminal primers is affected, because such variation of the binding site of an internal primer is visible in the sequence from the overlapping PCR product. If such a small deletion occurs within the analysed region, the probability that the critical terminal primers are affected is approximately 2*(d + p)/L which is very low for long exons. In addition, the terminal primers are usually located in intronic sequences and small deletions in these regions are rarely of any clinical relevance.
In contrast to the reliably detected small deletions, deletions longer than the PCR amplicons are never detected by sequencing because they cannot be amplified. Deletions of this size, affecting only part of a large exon, may not be detectable at all through MLPA when wide-spaced MLPA probes are used.
Most critical appear to be medium-sized deletions which can be detected by chance only when using wide-spaced MLPA probes. Sequencing fails to detect such deletions if binding of a terminal primer or both primers of adjacent amplicons in the overlapping region are affected. Therefore, deletions smaller than an amplicon but at least of the size of the region between primers in the overlap of amplicons are not detected with high sensitivity.
For an exon of length l that is divided into q amplicons with overlapping regions of a, defined by primer of constant lengths p, the likelihood P of missing a deletion of the size d (l/q > d > a-2p) which occurs at a random site in the analysed region can be calculated as:
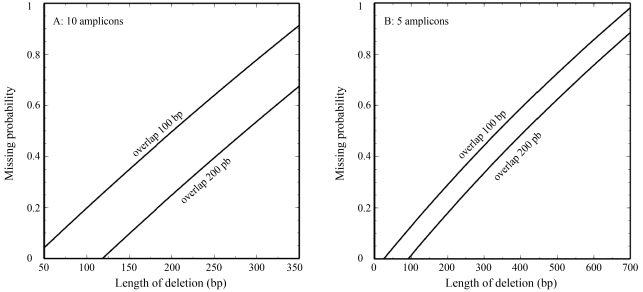
Figure 2A shows the function P(d) for parameters typical for the analysis of BRCA1 exon 11. Obviously, a substantial fraction of possible medium-sized deletions are not detectable by sequencing, but the chance of their detection can be increased by reducing the number of PCR amplicons and by expanding the overlapping regions (Figure 2).
Figure 2.
Probability of missing medium-sized deletions in long exons by sequencing of overlapping PCR products depends on the number of amplicons and the size of the overlapping regions. Based on a simplified model of deletions occurring with the same likelihood over the analysed region, and analysis with primers of constant (25 nt) lengths resulting in q amplicons of the same extent, calculations (see Results section) were performed for a region of the size of BRCA exon 11 (3426 bp). A: Sequence analysis based on 10 PCR amplicons overlapping 100 bp or 200 bp. B: Sequence analysis based on 5 PCR amplicons overlapping 100 bp or 200.
Initiated by the MLPA-based detection of deletion that was missed despite sequence analysis covering the affected region, we performed model calculations to estimate the fractions of medium-sized deletions that cannot be discovered by conventional methods. These were surprisingly high under typical conditions, especially for deletion sizes just below the amplicon lengths. The currently used BRCA1 MLPA kits contain only two probes for exon 11, and therefore only few of such medium-sized deletions can be found using this approach. We conclude that compared to small and large deletions, which are reliably found using conventional sequencing and MLPA, the sensitivity of detection of medium-sized deletions with these methods is very low. To increase sensitivity, a set of narrow-spaced MLPA probes could be used. Alternatively, amplification of complete large exons by long PCR followed by high resolution gel electrophoresis should resolve the problem.
Acknowledgments
We are grateful to Ms Astrid Deissler for arithmetic support of model calculations.
References
- Arnold N, Peper H, Bandick K, Kreikemeier M, Karow D, Teegen B, Jonat W. Establishing a control population to screen for the occurrence of nineteen unclassified variants in the BRCA1 gene by denaturing high-performance liquid chromatography. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;782:99–104. doi: 10.1016/s1570-0232(02)00696-7. [DOI] [PubMed] [Google Scholar]
- Hogervorst FB, Nederlof PM, Gille JJ, Mc Elgunn CJ, Grippeling M, Pruntel R, Regnerus R, van Welsem T, van Spaendok R, Menko FA, et al. Large genomic deletions and duplications in the BRCA1 gene identified by a novel quantitative method. Cancer Res. 2003;63:1449–1453. [PubMed] [Google Scholar]
- King MC, Marks JH, Mandell JB. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- van der Stoep N, van Paridon CD, Janssens T, Krenkova P, Stambergova A, Macek M, Matthijs G, Bakker E. Diagnostic guidelines for high-resolution melting curve (HRM) analysis: an interlaboratory validation of BRCA1 mutation scanning using the 96-well LightScanner. Hum Mutat. 2009;30:899–909. doi: 10.1002/humu.21004. [DOI] [PubMed] [Google Scholar]
- Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]