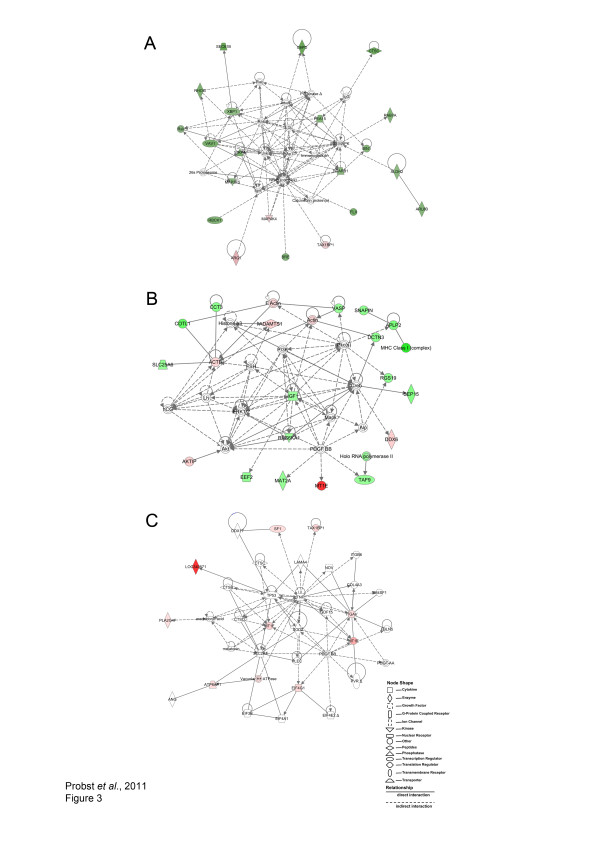
Figure 3.
Networks built using differentially expressed genes in L. amazonensis-infected and uninfected macrophages. C57BL/6 or CBA macrophages were cultured, infected and processed for microarray analysis as described in Materials and Methods. Considering the modulated genes in C57BL/6 infected macrophages, the immunological disease and cell morphology network (A), as well as the protein synthesis, cellular development and cell death network (B) were modeled by IPA®. Considering the modulated genes in CBA infected macrophages, the lipid metabolism, cellular movement, and small molecule biochemistry network was built by IPA® (C). C57BL/6 and CBA macrophages were cultured separately, then infected and processed for microarray analysis as described in Materials and Methods. Similar to Figure 2, the above networks are displayed as a series of nodes (genes or gene products) and edges (or lines, corresponding to biological relationships between nodes). Nodes are displayed using shapes as indicated in the key. Nodes marked in red were found to be highly expressed in infected macrophages. Nodes marked in green were found to be highly expressed in uninfected macrophages. Unmarked nodes were added by IPA® due to a high degree of probability of involvement in a given network. The node color intensity is an indication of the degree of up-(red) or down-(green) regulation of genes observed in the biological network analysis from both C57BL/6 and CBA macrophages in response to infection. Solid lines denote direct interactions, whereas dotted lines represent indirect interactions between the genes represented in this network.