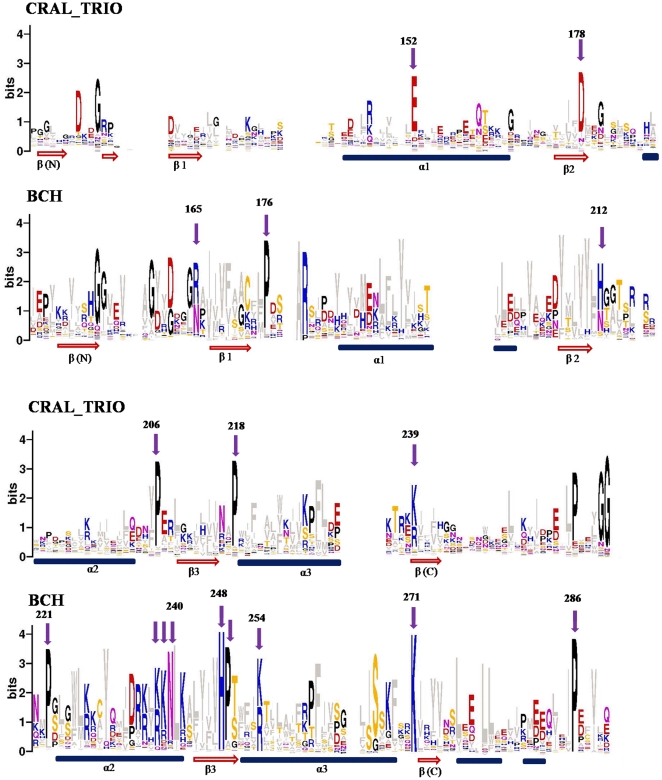
Figure 2. Sequence logos of CRAL_TRIO and BCH domains.
The sequence logos derived from 175 BCH and 78 CRAL_TRIO domain sequences are shown in this figure. The conserved residues are marked with arrows and the numbering is given according to the yeast Sec14p protein (NCBI accession: NP_013796) for CRAL_TRIO domains and the human BNIP-2 protein (NCBI accession: NP_004321) for BCH domains. The approximate positions of α-helices and β-beta strands are indicated at the bottom by blue cylinders and red arrows. In order to avoid any biased data, the ‘BCH-like’ groups (NF1 and RhoGEFs) were excluded from the logo calculation. These logos reveal characteristic differences between BCH and CRAL_TRIO domains. Unique positions within the two groups are marked by arrows. BCH domains have a unique signature motif R(R/K)h(R/K)(R/K)NL(R/K)xhhhhHPs in which ‘h’ refers to any large and hydrophobic residue and ‘s’ is a small and weekly polar residue (A, T, G, S). This motif is missing in CRAL_TRIO domains. The motif contains a patch of positively charged residues referred to as an Arg/Lys patch. Similarly, as exemplified by the aromatic residue in the middle of three α-helices, many of the hydrophobic residues (shown in grey) are conserved at various positions. The conservation of long and hydrophobic residues in the β-strands provides a hydrophobic surface.