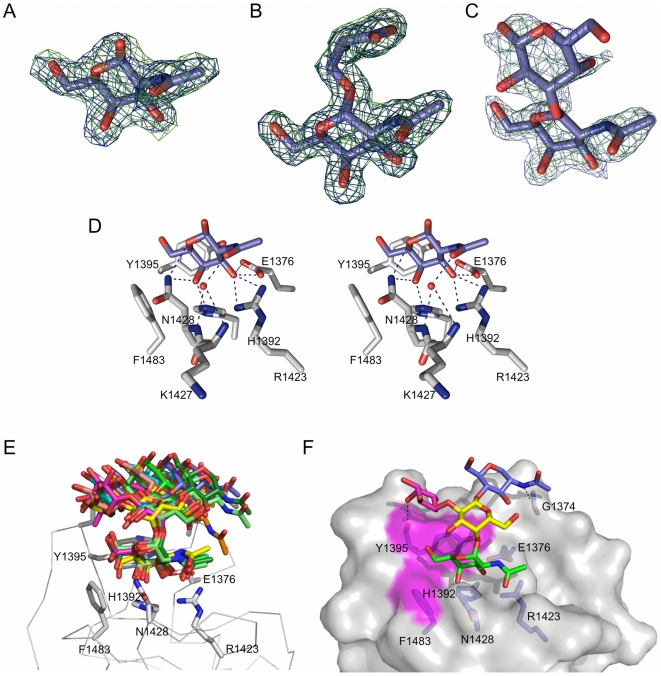
Figure 4. Structural analysis of CBM32-5 with additional carbohydrates.
(A) Electron density for GalNAc shown as maximum-likelihood/σA [59] -weighted 2F obs-F calc maps contoured at 1 σ (both maps at 0.31 e−/Å3) produced by refinements prior to modeling the sugar (green) and with the sugar included (blue). (B) Electron density for serinyl-Tn antigen shown as maximum-likelihood/σA [59] -weighted 2F obs-F calc maps contoured at 1 σ (both maps at 0.45 e−/Å3) produced by refinements prior to modeling the sugar (green) and with the sugar included (blue). (C) Electron density for GalNAc-β-1,3-galactose shown as maximum-likelihood/σA [59] -weighted 2F obs-F calc maps contoured at 0.8 σ (both maps at 0.34 e−/Å3) produced by refinements prior to modeling the sugar (green) and with the sugar included (blue). (D) Divergent stereo view of the key interactions between the binding site of CBM32-5 and GalNAc. This also represents the mode of interaction between the CBM and the serinyl-Tn antigen and GalNAc-β-1,3-galactose, which all have identical hydrogen bonding patters. Hydrogen bonds are shown as dashed black lines. (E) Models of CBM32-5 in complex with GalNAc-α-1,4(Fuc-α-1,2)-Gal-β-1,4-GlcNAc-OMe produced by molecular dynamics simulations. An ensemble of ten energy minimized models is given with each model representing a group of energetically similar models. Relevant residues in the binding site are shown as grey sticks with the backbone of the protein shown as a Cα-ribbon. (F) A surface representation of the lowest energy model of CBM32-5 bound to the tetrasaccharide (GalNAc is shown in green, fucose in pink, galactose in yellow, and GlcNAc in blue). The surfaces contributed by Y1395 and F1483 are shown in magenta and additional hydrogen bonds made outside of the primary galactose binding site shown as dashed lines.