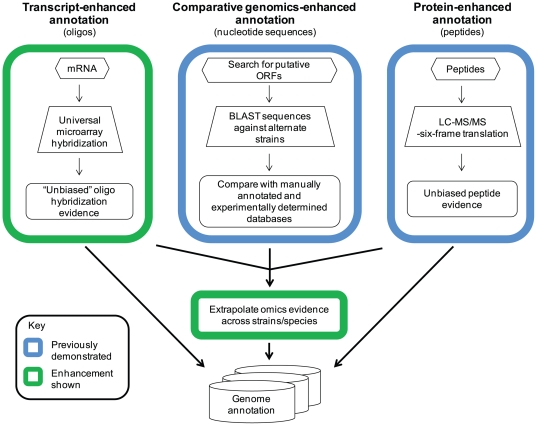
Figure 1. Schematic showing the comparative omics-based genome annotation workflow employed for annotation refinement.
Transcriptomic data generated from an unbiased universal Yersinia microarray and peptide data matched to a 6-frame genome translation were layered on existing genome annotations to validate predicted protein coding sequences and identify annotation anomalies. This evidence can be used independently or combined with putative protein identifications derived from a comparative genomics approach for genome annotation refinement.