Abstract
Background
HIV-1 replication depends on a delicate balance between cellular co-factors and antiviral restriction factors. Lens epithelium-derived growth factor (LEDGF/p75) benefits HIV, whereas apolipoprotein B mRNA-editing catalytic polypeptide-like 3G (APOBEC3G), tripartite motif 5alpha (TRIM5α), and tetherin exert anti-HIV activity. Expression levels of these proteins possibly contribute to HIV-1 resistance in HIV-1-exposed populations.
Methodology/Principal Findings
We used real-time PCR and flow cytometry to study mRNA and protein levels respectively in PBMC and PBMC subsets. We observed significantly reduced LEDGF/p75 protein levels in CD4+ lymphocytes of HIV-1-exposed seronegative subjects relative to healthy controls, whereas we found no differences in APOBEC3G, TRIM5α, or tetherin expression. Untreated HIV-1-infected patients generally expressed higher mRNA and protein levels than healthy controls. Increased tetherin levels, in particular, correlated with markers of disease progression: directly with the viral load and T cell activation and inversely with the CD4 count.
Conclusions/Significance
Our data suggest that reduced LEDGF/p75 levels may play a role in resistance to HIV-1 infection, while increased tetherin levels could be a marker of advanced HIV disease. Host factors that influence HIV-1 infection and disease could be important targets for new antiviral therapies.
Introduction
Human immunodeficiency virus type 1 (HIV-1) interacts with many cellular host proteins during its replication cycle [1]–[3]. Some of these proteins are required for HIV-1 replication while others exhibit antiviral activity. Lens epithelium-derived growth factor p75 (LEDGF/p75) [4] is a cellular co-factor, whereas apolipoprotein B mRNA-editing catalytic polypeptide-like 3G (APOBEC3G, A3G) [5], tripartite motif 5alpha (TRIM5α) [6], and tetherin (BST2, CD317, HM1.24) [7] are cellular factors with distinct antiviral activities. The main working mechanisms of these factors can be summarized as follows. LEDGF/p75 assists in the integration of viral cDNA into specific regions of the host's genetic material [8], [9]. APOBEC3G inhibits HIV replication by inducing G-to-A hypermutation of the viral HIV-1 genome during reverse transcription [5], [10]. TRIM5α structurally disorders the retroviral capsid, leading to the interruption of the natural “uncoating” process in a species-specific manner [11], [12]; and was recently described to promote innate immune signaling [13]. Tetherin, finally, inhibits HIV release by tethering newly formed retrovirus particles to the cell membrane [7].
The significance of these four proteins as possible correlates of protection against HIV-1 remains to be determined. Several studies on peripheral blood mononuclear cells (PBMC) have found that APOBEC3G mRNA levels correlate positively with the CD4 count and negatively with the viral load of untreated HIV-1-infected subjects [14]–[16], suggesting that APOBEC3G contributes to the control of HIV-1 infection. However, other studies did not find such correlations for APOBEC3G [17]–[19] or TRIM5α [20] mRNA. In addition, certain studies compared the expression of APOBEC3G or TRIM5α between distinct study populations like untreated HIV-1-infected subjects, healthy controls, and HIV-1-exposed seronegative individuals. Some studies observed lower mRNA expression levels of APOBEC3G [14], [17], [19] or TRIM5α [20] in untreated HIV-1-infected subjects compared to healthy controls, although the opposite was also found for APOBEC3G [15]. APOBEC3G mRNA and protein expression levels were also studied respectively in PBMC and PBMC-subsets (CD4+ and CD8+ lymphocytes, and CD14+ monocytes) of HIV-1-exposed seronegative subjects relative to healthy controls [16], [19], [21]. Here too, conflicting data were obtained, showing either similar [19] or higher [16], [21] levels of antiviral APOBEC3G in HIV-1-exposed seronegative subjects relative to healthy controls.
In the present study, we investigated mRNA and protein expression levels of the 4 HIV-1-related factors LEDGF/p75, APOBEC3G, TRIM5α, and tetherin in HIV-1-exposed seronegative subjects, healthy controls, untreated HIV-1-infected subjects, and antiretroviral therapy-treated HIV-1-infected subjects. HIV-1-exposed seronegative subjects are individuals who remain HIV-1 seronegative despite frequent and unprotected exposure to HIV, and are observed in cohorts of commercial sex workers, men having sex with men, intravenous drug users, and discordant couples [22]. In this study, HIV-1-exposed seronegative subjects were enrolled from a Senegalese cohort of HIV-1 serodiscordant couples. Real-time quantitative polymerase chain reaction (qPCR) was applied to study mRNA expression in PBMC. Protein expression levels were determined simultaneously in different PBMC subsets by flow cytometry. We hypothesized that expression levels of the above-mentioned proteins may contribute to the putative in vivo HIV-1 restriction in HIV-1-exposed seronegative subjects.
Materials and Methods
Ethics Statement
The study was approved by the Internal Review Board of the Institute of Tropical Medicine (Antwerp, Belgium) and by the Ethical Committees of the Senegalese Ministry of Health (Dakar, Senegal) and the University Hospital of Antwerp (Belgium). All study subjects gave written informed consent prior to enrollment.
Study population
This study was conducted on a cohort of heterosexual couples recruited at the Department of Infectious Diseases at the Fann University Teaching Hospital, Dakar, Senegal, as previously described [23]. Twenty three HIV-1 exposed seronegative subjects (HESN) in HIV-1 serodiscordant couples, 23 healthy controls (HC) in HIV-1-negative seroconcordant monogamous couples, and 45 HIV-1-infected patients in HIV-1 serodiscordant or HIV-1-positive seroconcordant couples were studied. Within the group of 45 HIV-1-infected patients, 9 patients were untreated HIV-1-infected subjects (HIV-UT) and 36 patients were antiretroviral therapy-treated HIV-1-infected subjects (HIV-ART). All couples had a sexual relationship of at least 7.5 years. HESN and HC were matched for gender. Blood samples and standard questionnaires with information on socio-demographics and sexual behavior were collected. Male condom provision and sexual risk reduction counseling were provided during every visit.
Sample collection and processing
Whole blood samples were collected in EDTA tubes. CD4+ T cell counts and T cell activation levels were determined in fresh whole blood using CD3, CD4, CD8, and CD38 fluorochrome-labeled antibodies and a FACSCalibur flow cytometer (BD Biosciences) like previously reported [23]. In this article, the T cell activation status is determined by the percentage of CD38+ cells among the CD8+ T cells. Plasma and PBMC were separated from fresh whole blood, frozen down at −80°C and in liquid nitrogen, respectively, and shipped to Belgium. HIV and HSV-2 (Herpes simplex virus type 2) status were determined in plasma by ELISA. HIV-1 viral loads were quantified in plasma by the Amplicor HIV-1 Monitor assay, version 1.5 (Roche Diagnostics GmbH). PBMC were thawed, washed twice, and counted prior to mRNA and protein expression analysis.
RNA extraction, DNase treatment and reverse transcription
PBMC samples were dissolved in TRIzol® Reagent (Invitrogen) and stored at −80°C for less than three months. Total RNA was isolated according to the manufacturer's instructions (Invitrogen). Removal of genomic DNA was performed with the DNA-free Kit (Ambion) following the manufacturer's protocol. Additional wash steps, once with 200 µl isopropanol (in the presence of 0.7 µl of glycogen) and twice with 75% ethanol, were performed to remove traces of contaminants. The NanoDrop ND-1000 spectrophotometer (Thermo Scientific) was used to quantify total RNA levels. Total DNase-treated RNA was stored at −80°C for less than one month. The iScript cDNA synthesis Kit (Bio-Rad Laboratories NV-SA) was applied to 400 ng of total DNase-treated RNA following the manufacturer's instructions. In parallel, 100 ng of the same RNA was dissolved in RNase/DNase-free water (no-RT control) at the same final concentration (20 ng/10.5 µl) as the RT (reverse transcribed) samples. Both RT and no-RT samples were stored at −80°C for less than one month. For each sample, the difference in quantification cycle (Cq)-value between RT and no-RT samples was higher than 10. During optimization of the qPCR protocol, RNA quality was evaluated by RNA gel electrophoresis (1% agarose gel). The 18S and 28S fragments were visualized with GelRed (VWR) versus a 1-kb DNA ladder (Invitrogen) and were observed in the expected 1∶2 ratio on representative samples.
Real-time PCR
Messenger RNA (mRNA) expression levels were quantified by real-time PCR using the Bio-Rad CFX96 system and SYBR Green mix (Bio-Rad). Target-specific primers were designed using the Harvard website: http://pga.mgh.harvard.edu/primerbank/, except for TRIM5α and TBP, for which Primer3 software (v.0.4.0) was used. Specificity was tested using BLAST (NCBI). All primer pairs, except for TRIM5α, could be designed to span at least one intron. Name, GeneID, primer sequences, exon localization and amplicon length are listed in Table 1. The primer mixes were prepared by combining Forward (Fw) and Reverse (Re) Primers (Eurogentec), both at a concentration of 2 pmol/µl. Each optimized 25 µl reaction contained 12.5 µl of iQ SYBR Green Supermix (Bio-Rad), 2 µl of the primer mix and 5, 10 or 20 ng of the cDNA template at a volume of 10.5 µl. Each assay included, in triplicate, a standard curve, a no-template control (water), the RT samples, and, in the case of TRIM5α, the no-RT samples. The following subsequent heating steps were included as PCR cycling conditions: 50°C (10 min), 95°C (3 min), 40 cycles of 95°C (10 s) and 60°C (30 s) with fluorescence capturing following each cycle, 95°C (10 s), and melting curve acquisition with fluorescence capturing every 5 s during a temperature increase in 0.5°C increments from 65°C to 95°C. The amplicon length of the PCR product was validated by 2% agarose DNA gelelectrophoresis. PCR products were visualized with GelRed (VWR) versus a 100-bp DNA ladder (Invitrogen).
Table 1. Primers used for reverse transcriptase quantitative polymerase chain reaction (RT-qPCR).
| GeneID | Primer Sequence 5′ to 3′ | Exon localization | Amplicon length | ||
| Genes of interest | |||||
| LEDGF/p75 | 11168 | Fw*Re† | TCGACTTCAAAGGATACATGCTG GAGCTTGTTGCATTGTGACCT | 1213 | 122 |
| APOBEC3G | 60489 | FwRe | TCAGAGGACGGCATGAGACTT TGGAGCCTGGTTGCATAGAAA | 55 and 6 | 107 |
| TRIM5alpha | 85363 | FwRe | TGCTGGCTTCCAACCTGAT ACAGAGAGGGGCACAATGAA | 88 | 165 |
| Tetherin (BST2) | 684 | FwRe | GAGTGTCGCAATGTCACCCAT GGAAGCCATTAGGGCCATCAC | 11 and 2 | 120 |
| Reference genes | |||||
| UBC‡ | 7316 | FwRe | ATTTGGGTCGCAGTTCTTG TGCCTTGACATTCTCGATGGT | 12 | 123 |
| B2M§ | 567 | FwRe | GGCTATCCAGCGTACTCCAAA CGGCAGGCATACTCATCTTTTT | 1 and 22 | 246 |
| GAPDH∥ | 2597 | FwRe | TGTTGCCATCAATGACCCCTT CTCCACGACGTACTCAGCG | 35 | 202 |
| RPL13A¶ | 23521 | FwRe | CGAGGTTGGCTGGAAGTACC CTTCTCGGCCTGTTTCCGTAG | 78 | 121 |
| TBP# | 6908 | FwRe | ACCCAGCAGCATCACTGTTT CCAAGCCCTGAGCGTAAG | 1 and 22 | 200 |
Forward primer,
Reverse primer,
Ubiquitin C,
Beta-2-Microglobulin,
Glyceraldehyde 3-phosphate dehydrogenase,
60S ribosomal protein L13a, and
TATA-binding protein.
mRNA data analysis
The obtained results (Cq-values) were exported from the Bio-Rad CFX Manager version 1.5 and imported into qbasePLUS software, version 1.5, from BioGazelle (Ghent, Belgium) following baseline subtraction and manual threshold matching. The GeNorm applet selected the reference genes B2M and GAPDH out of five genes (UBC, B2M, GAPDH, RPL13A, TBP) as the most stably expressed across the different study populations (25 samples, all applied in triplicate). Both reference genes were used to normalize target gene expression in the qbasePLUS software. For the standard curve, a 2-fold serial dilution of cDNA of one Caucasian healthy control was used. Cq-values were converted into relative expression values taking into account amplification efficiencies, inter-run variations, and normalization factors. Outliers were excluded when a difference of more than 1.0 was registered in the Cq-values. Finally, CNRQ (Calibrated Normalized Relative Quantity) values were exported from the qbasePLUS software and statistically investigated.
Intracellular and surface protein staining
The protein expression levels of LEDGF/p75, APOBEC3G, TRIM5α, and tetherin were investigated in CD4+ T cell and CD14+ monocyte subsets based on simultaneous intracellular and/or cell surface staining of PBMC. The following reagents were used: 0.1% bovine serum albumin (Acros Organics) and 0.05% sodium azide (Merck) in phosphate buffered saline (PBS, Lonza) as washing buffer; Reagents A and B to fix and permeabilize cells, respectively (Leucoperm kit, AbD Serotec); primary mouse unlabeled antibody (isotype IgG1 and IgG2a) and anti-LEDGF/p75 (IgG1) from BD Biosciences, anti-APOBEC3G and anti-TRIM5α (both IgG1) from the AIDS Research and Reference Reagent Program (Division of AIDS, NIAID, NIH), and anti-tetherin (IgG2a) as kindly provided by Chugai Pharmaceutical Co., Japan; secondary FITC-labeled antibody (BD Biosciences); normal mouse serum (eBioscience); membrane antibodies anti-CD4-PerCP, anti-CD45RO-APC, anti-CD38-PE, anti-CD14-APC, and anti-CD16-PE from BD Biosciences; and 1% paraformaldehyde (Sigma-Aldrich). LEDGF/p75, APOBEC3G, TRIM5α, and tetherin were analyzed using an in-house optimized intracellular staining (ICS) protocol [24], [25]. Cells were washed (630×g, 10 min, RT), fixed (10 min, RT), washed twice, permeabilized (30 min, 4°C), washed, incubated with primary antibody (24 h, 4°C), washed twice, incubated with secondary antibody supplemented with reagent B (30 min, 4°C), washed twice, incubated with normal mouse serum (10 min, 4°C), incubated with a membrane antibody cocktail (CD4/CD45RO/CD38 or CD4/CD14/CD16 to study lymphocytes or monocytes, respectively, 20 min, 4°C), washed twice, and dissolved in 1% paraformaldehyde prior to data acquisition (FACSCalibur flow cytometer, BD Biosciences). To study membrane tetherin levels, PBMC were processed according to the protocol above with a 30-min anti-tetherin incubation at 4°C, without fixation or permeabilization steps. In general, at least 25000 and 50000 events were respectively measured to study expression patterns in CD4+ lymphocytes and CD14+ monocytes. Data analysis with CellQuest Pro Software (BD Biosciences) produced median fluorescence intensity (MFI) values, reflecting the median amount of bound antibody per cell and thus, the protein's abundance. Host protein MFI-values were corrected for non-specific background staining by subtracting isotype MFI-values.
Statistical analyses
Differences in expression levels between individuals of different study groups were investigated with non-parametric Mann-Whitney U tests. Correlation analyses were performed using non-parametric Spearman's rank correlation tests. Observations were considered statistically significant when p<0.05. Statistical analyses were performed with SPSS version 17.0, and graphs were drawn with GraphPad Prism, version 5.
Results
Study population
Twenty three HIV-1-exposed seronegative subjects (HESN), 23 healthy controls (HC), and 45 HIV-1-infected patients were studied. Nine HIV-1-infected patients were untreated (HIV-UT) and 36 were antiretroviral therapy-treated (HIV-ART). Several parameters like age, gender, duration of the sexual relationship, CD4 count, number of sexual contacts per month, condom use, and HSV-2 serostatus were compared between HESN and HC, HIV-UT and HC, and HIV-ART and HIV-UT (Table 2). The HC subjects were in a monogamous relationship for at least 7.5 years at the time of enrollment. They reported a significantly lower percentage of condom use. HIV-1-infected subjects were more often male individuals. Therapy-naïve HIV-1 infected subjects (HIV-UT) showed significantly lower CD4 count levels than HC, and a higher prevalence of HSV-2 seropositivity.
Table 2. Characteristics of HIV-exposed seronegative, HIV-1-infected, and healthy control subjects included in the study.
| HC*(n = 23) | HESN†(n = 23) | HIV-UT‡(n = 9) | HIV-ART§(n = 36) | p-value | |||
| HESN vs. HC | HIV-UT vs. HC | HIV-ART vs. HIV-UT | |||||
| Age, years | 43(37–46) | 41(33–47) | 46(37–51) | 44(36–51) | 0.409 | 0.449 | 0.955 |
| Gender (% male) | 47.8 | 43.5 | 88.9 | 72.2 | 0.770 | 0.036 | 0.303 |
| CD4 count (cells/µl) | 789(703–1090) | 738(548–891) | 228(133–337) | 334(195–412) | 0.215 | <0.001 | 0.294 |
| Number of sexual contacts per month | 8(3–12) | 5(4–8) | 5(4–11) | 7(4–8) | 0.164 | 0.831 | 0.831 |
| Duration of sexual relation, years | 12(8–19) | 11(8–18) | 11(9–18) | 10(8–16) | 0.938 | 0.866 | 0.540 |
| Condom use (% always) | 0 | 61 | 44 | 50 | <0.001 | 0.001 | 0.768 |
| HSV-2 serostatus (% HSV-2 positive) | 22 | 45 | 78 | 59 | 0.095 | 0.004 | 0.301 |
Data are median (interquartile range) values or percentages if indicated.
Healthy controls,
HIV-exposed seronegatives,
therapy-naïve HIV-1-infected subjects,
antiretroviral therapy-treated HIV-1-infected subjects. P-values (Mann Whitney U) below 0.05 are in bold.
Differences in mRNA expression in PBMC
The mRNA expression levels of LEDGF/p75, APOBEC3G, TRIM5α, and tetherin were analyzed for all study subjects in PBMC by real-time qPCR (Table 3). None of the genes displayed a significant difference in mRNA expression between HESN and HC. HIV-UT showed significantly lower levels of LEDGF/p75 (p = 0.008) and higher levels of tetherin (p = 0.027) than HC. Additionally, higher expression levels of APOBEC3G (p = 0.078), TRIM5α (p = 0.016), and tetherin (p<0.001) were observed in HIV-UT relative to HIV-ART subjects.
Table 3. Expression levels of LEDGF/p75, APOBEC3G, TRIM5α, and tetherin in PBMC of HIV-exposed seronegative, HIV-infected, and healthy control subjects.
| HC* | HESN† | HIV-UT‡ | HIV-ART§ | p-value | |||
| HESN vs. HC | HIV-UT vs. HC | HIV-ART vs. HIV-UT | |||||
| mRNA expression | N = 20 | N = 20 | N = 9 | N = 31 | |||
| LEDGF/p75 | 1.22(0.88–1.50) | 1.03(0.83–1.36) | 0.72(0.53–1.11) | 0.92(0.70–1.10) | 0.387 | 0.008 | 0.391 |
| APOBEC3G | 0.98(0.79–1.23) | 0.99(0.86–1.25) | 1.29(1.00–1.38) | 0.96(0.83–1.22) | 1 | 0.172 | 0.078 |
| TRIM5α | 1.08(0.79–1.42) | 0.85(0.74–1.27) | 1.22(0.92–1.89) | 0.78(0.65–1.18) | 0.372 | 0.370 | 0.016 |
| Tetherin | 1.12(0.88–1.36) | 1.10(0.96–1.29) | 1.46(1.14–1.85) | 0.99(0.78–1.06) | 1 | 0.027 | <0.001 |
| Protein expression | N = 23 | N = 23 | N = 9 | N = 36 | |||
| CD4+ lymphocytes | |||||||
| LEDGF/p75 | 113.10(97.62–126.05) | 98.80(77.95–110.13) | 121.91(85.58–53.40) | 106.85(94.77–122.80) | 0.022 | 0.516 | 0.371 |
| APOBEC3G | 39.61(35.48–42.93) | 38.01(31.08–44.88) | 39.68(36.58–47.88) | 40.64(35.53–47.08) | 0.613 | 0.572 | 0.691 |
| TRIM5α | 107.57(96.87–123.01) | 112.80(93.67–125.22) | 126.13(100.47–156.08) | 115.15(100.26–134.41) | 0.939 | 0.148 | 0.427 |
| Membrane tetherin | 4.16(3.71–4.68) | 4.35(3.94–4.81) | 6.55(5.34–9.29) | 4.77(4.03–5.19) | 0.733 | <0.001 | <0.001 |
| Total tetherin | 5.32(4.64–6.74) | 5.33(4.43–6.54) | 10.73(7.20–18.52) | 5.88(5.43–6.90) | 0.809 | 0.003 | 0.004 |
| CD14+ monocytes | |||||||
| LEDGF/p75 | 49.26(40.55–70.39) | 45.51(36.73–71.27) | 77.05(62.22–86.85) | 45.65(40.94–59.45) | 0.750 | 0.005 | 0.002 |
| APOBEC3G | 102.09(81.37–128.12) | 98.62(80.19–13.40) | 123.28(111.52–172.55) | 105.30(90.89–116.28) | 0.328 | 0.031 | 0.002 |
| TRIM5α | 280.38(234.04–335.57) | 283.79(235.40–332.42) | 398.98(317.44–513.08) | 287.94(257.72–331.20) | 0.869 | 0.005 | 0.002 |
| Membrane tetherin | 1.62(0.00–6.67) | 2.42(0.00–4.95) | 11.77(6.38–21.86) | 4.78(1.19–7.17) | 0.894 | 0.002 | 0.005 |
| Total tetherin | 49.28(40.00–65.42) | 49.84(40.99–55.70) | 79.48(59.27–113.97) | 46.13(38.27–50.87) | 0.956 | 0.003 | <0.001 |
Data are median (interquartile range) values depicting Calibrated Normalized Relative Quantity (CNRQ-)values for mRNA and Median Fluorescence Intensity (MFI-)values for protein expression.
Healthy controls,
HIV-exposed seronegatives,
therapy-naïve HIV-1-infected subjects,
antiretroviral therapy-treated HIV-1-infected subjects. P-values (Mann Whitney U) below 0.05 are in bold. Of note, due to sample loss during RNA extraction, data on mRNA expression were gathered for slightly less study subjects.
Differences in protein expression in lymphocytes and monocytes
Next, we analyzed protein expression levels of LEDGF/p75, APOBEC3G, TRIM5α, and tetherin in CD4+ lymphocytes and CD14+ monocytes by a novel intracellular staining assay [25]. Expression of LEDGF/p75 was found to be significantly reduced (p = 0.022) in CD4+ lymphocytes of HESN compared to HC (Table 3). APOBEC3G, TRIM5α, and tetherin protein levels were similar for HESN and HC. HIV-UT showed significantly higher tetherin levels in CD4+ lymphocytes and significantly higher levels of all four proteins in CD14+ monocytes compared to HC and HIV-ART.
Correlations between mRNA and protein levels
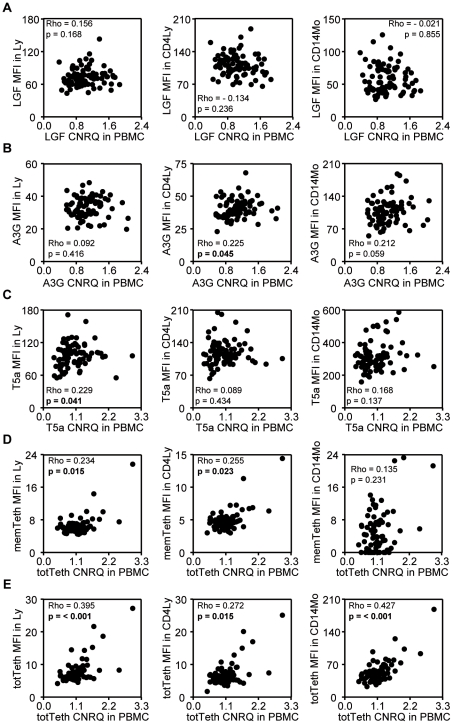
Subsequently, we correlated mRNA levels in PBMC with protein levels specifically in lymphocytes and monocytes within the entire study population including HESN, HC, HIV-UT, and HIV-ART subjects (Figure 1). For most HIV-related host factors, we observed a significant positive correlation between mRNA and protein level as was most prominent for total tetherin, whereas no significant correlations were observed for LEDGF/p75.
Figure 1. Correlations between mRNA and protein levels.
Correlation patterns were studied in all study subjects between mRNA (CNRQ-values, x-axis) and protein (MFI-values, y-axis) levels of (A) LEDGF/p75 (LGF), (B) APOBEC3G (A3G), (C) TRIM5α (T5a), (D) membrane tetherin (memTeth), and (E) total tetherin (totTeth). mRNA levels were studied in PBMC while protein levels were analyzed in lymphocytes (Ly, left), CD4+ lymphocytes (CD4Ly, middle), and CD14+ monocytes (CD14Mo, right). CNRQ-values refer to Calibrated Normalized Relative Quantity of mRNA expression levels and MFI-values refer to Median Fluorescence Intensity of protein expression levels. Spearman's Rho (Rho) and p-values were obtained by non-parametric Spearman's rank correlation tests. P-values below 0.05 are in bold.
Correlates of reduced LEDGF/p75 in HESN
To obtain insight into the possible role played by LEDGF/p75 in resistance to HIV-1 infection, we studied the reduced LEDGF/p75 expression levels in HESN considering the duration of the sexual relation, the number of sexual contacts per month, the frequency of condom use, and the HSV-2 serostatus (Table 4). We found that LEDGF/p75 expression levels were most reduced among HESN with the highest number of sexual contacts per month (p = 0.065), whereas no difference in LEDGF/p75 expression was found in the HIV-1-negative control group (Table 4).
Table 4. Expression levels of LEDGF/p75 versus HIV-1 risk/exposure parameters within different study populations of interest.
| HC* | HESN† | |||||
| subgroup 1a | subgroup 2b | P-value | subgroup 1 | subgroup 2 | P-value | |
| Sexual contacts/month(<a vs ≥b median value) | 109.47(98.59–124.73) | 114.26(94.84–137.76) | 0.537 | 105.63(95.04–112.09) | 85.37(74.88–104.96) | 0.065 |
| Duration of relation, years(<a vs ≥b median value) | 113.10(99.57–141.30) | 110.90(89.88–124.04) | 0.284 | 104.58(73.50–111.30) | 95.46(81.66–112.71) | 1.000 |
| Condom use(alwaysa vs not alwaysb) | - | 110.23(96.24–126.13) | ND | 98.30(69.33–111.37) | 98.80(81.66–112.66) | 0.571 |
| HSV-2 status(negativea vs positiveb) | 110.23(95.70–124.73) | 123.96(96.80–132.58) | 0.502 | 98.30(76.20–110.11) | 101.50(85.31–126.40) | 0.468 |
Healthy controls,
HIV-exposed seronegatives. Each HIV-1 risk/exposure parameter (left column) was used to split the study population in two subgroups (a subgroup 1 versus b subgroup 2) based on a categorical or on a numerical parameter. Numerical parameters use median values (see Table 2) to split the study group in two subgroups comparable in size. LEDGF/p75 expression levels are median (interquartile range) values. Within each study population, the P-value (Mann Whitney U) was calculated for each HIV-1 risk/exposure parameter. ND: not determined as none of the subjects used a condom.
Correlations between expression levels and T cell activation
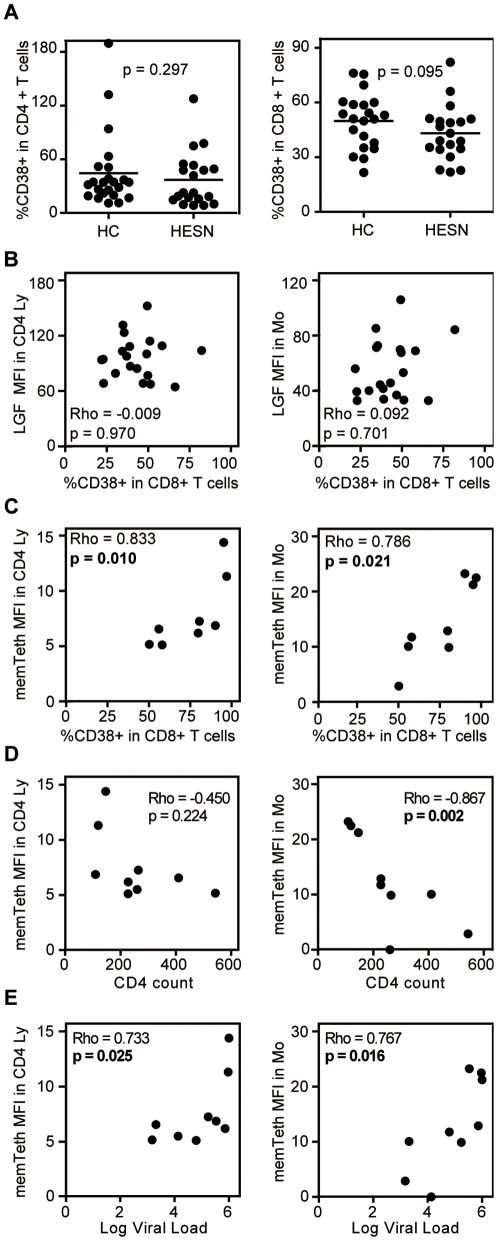
Finally, we investigated whether the observed differences in mRNA and/or protein expression level could be correlated to T cell activation levels and/or disease status (viral load and CD4 count). Whereas no significant difference was observed in CD4+ T cell activation level (% CD38+ cells among the CD4+ T cells, p = 0.297), HESN showed a trend towards lower CD8+ T cell activation levels than HC (p = 0.095) in Figure 2A. Reduced T cell activation levels among HESN did not correlate with their reduced LEDGF/p75 protein expression levels (p = 0.970, Figure 2B). Neither did we observe differences in LEDGF/p75 expression between distinct CD4+ lymphocytes based on CD45RO (memory) or CD38 (activation) staining among HESN (data not shown). In HIV-UT, T cell activation levels correlated directly with TRIM5α (p = 0.002) and tetherin (p = 0.086) mRNA levels, and inversely with LEDGF/p75 mRNA levels (p = 0.037) (data not shown). Membrane and total expression levels of tetherin, in particular, correlated directly with T cell activation and the viral load, and inversely with the CD4 count. These correlations were most prominent for membrane tetherin as is depicted in Figure 2C–E.
Figure 2. Statistical analyses within HESN and untreated HIV-1-infected subjects.
(A) Difference in CD4 (left) and CD8 (right) T cell activation status (y-axis) between HESN and HC (x-axis). P-values were obtained through the Mann Whitney U test. (B) Correlation pattern in HESN subjects between LEDGF/p75 protein levels (LGF, y-axis) in CD4+ lymphocytes (CD4 Ly, left) and CD14+ monocytes (Mo, right) and T cell activation status (x-axis). (C–E) The graphs depict correlation patterns in CD4+ lymphocytes (CD4 Ly, left) and CD14+ monocytes (Mo, right) of HIV-UT subjects between membrane tetherin protein levels (memTeth, y-axis) and T cell activation status (x-axis, C), CD4 count (cells/mm3) (x-axis, D) and log viral load (x-axis, E). The subject's general T cell activation status is described by the percentage of CD38+ cells among the CD8+ T cells (B,C). Spearman's Rho (Rho) and p-values were obtained by non-parametric Spearman's rank correlation tests. P-values below 0.05 are in bold. MFI = Median Fluorescence Intensity.
Discussion
In this study, we explored whether the expression of LEDGF/p75, APOBEC3G, TRIM5α, and tetherin could play a role in HIV-1 resistance. We studied mRNA levels in PBMC and protein levels in CD4+ lymphocyte and monocyte subsets. We observed significantly reduced levels of LEDGF/p75 protein expression in the CD4+ T cells of HESN compared to those of HC, whereas similar mRNA levels were found for both HESN and HC. Our findings of lower levels of LEDGF/p75 in CD4+ lymphocytes of HESN are in line with two recent HESN-studies showing reduced gene expression levels of several host genes important for HIV replication [26], [27]. Lower LEDGF/p75 protein levels may cause HIV-1 integration to occur less efficiently, thus contributing to HIV-1 resistance in HESN subjects. This hypothesis is supported by an in vitro experiment that found a direct correlation between LEDGF/p75 expression levels and the efficiency of HIV-1 integration in the host genome [28]. Other studies, however, reported that efficient knockdown or knock-out of LEDGF/p75 is not sufficient to completely abolish HIV-1 replication [29]–[31], suggesting that other factors may be needed in concert with LEDGF/p75 to limit HIV integration in HESN subjects.
We observed no differences in mRNA or protein levels of APOBEC3G, TRIM5α, or tetherin between HESN and HC. These findings are at variance with previous reports showing elevated APOBEC3G expression levels in HESN compared with HC [16], [21]. On the other hand, it has been suggested that reduced exposure to HIV-1 results in comparable APOBEC3G mRNA levels for HESN and HC over time [16]. In our HESN population, most HIV-1-infected partners were treated with antiretroviral therapy, and all couples received counseling to promote condom use. Therefore, the lack of distinct expression patterns may indeed result from the relatively low levels of HIV exposure in our population. Moreover, Reddy et al [19] observed similar levels of APOBEC3G mRNA between pre-infection values of seroconverters and exposed persistently negative individuals, suggesting that APOBEC3G mRNA levels would not contribute to protection against HIV-1.
Untreated HIV-1-infected patients (HIV-UT) showed significantly lower LEDGF/p75 and higher tetherin mRNA levels than HC, and higher APOBEC3G, TRIM5α, and tetherin mRNA levels than HIV-ART. At protein level, HIV-UT expressed significantly enhanced levels of tetherin in the lymphocytes and of all four proteins in the monocytes. These data suggest that HIV-1-infected subjects try to curb HIV-1 replication by increasing the expression level of the antiviral restriction factors.
Interestingly, increased levels of tetherin in HIV-UT correlated directly with T cell activation and the viral load and inversely with the CD4 count. This finding suggests that tetherin is not capable of controlling viral replication; instead it appears to be a marker of advanced HIV disease in therapy-naïve HIV-1-infected patients. Recently, Coleman et al [32] also reported increased tetherin expression levels upon exposure to HIV-1 without it being capable of restricting cell-to-cell spread of the virus. At least two other studies support our observations by suggesting that the tetherin-induced accumulation of unreleased virions at the cell membrane could actually promote cell-to-cell spread, e.g., by enhancing fusogenicity or regulating the integrity of the viral synapse [33], [34]. In turn, accumulation of virus particles at the cell membrane may trigger further increases in immune activation [35], [36]. Thus, tetherin may have a dual role during HIV-1 infection and disease. As an innate mediator, tetherin may be involved in the control of early or acute virus replication but it could become ineffective as the infection progresses.
T-cell activation levels also correlated directly with mRNA levels of TRIM5α but not with those of APOBEC3G; nevertheless we expect that similar mechanisms like those observed for tetherin are involved here. Indeed, type I interferons which are induced upon viral replication are known to upregulate APOBEC3G [15], [37]–[39], TRIM5α [40], [41], and tetherin [36], [42]. High-level interferon receptor expression on monocytes [21] may explain why our data showed relatively higher increases of APOBEC3G, TRIM5α, and tetherin in monocytes than in lymphocytes.
Intriguingly, HIV-UT expressed reduced mRNA levels in PBMC and elevated protein levels in monocytes for LEDGF/p75, relative to HC. The mRNA levels correlated inversely (p = 0.05; Rho = −0.667) and the protein levels tended to correlate directly (p = 0.170; Rho = 0.5) with the viral load of these patients (data not shown). These correlations suggest that LEDGF/p75 expression may also account for disease progression in HIV-UT, supporting recently described observations by Madlala et al [27]. Parallel to type I interferon induction, viral replication induces the production of several cytokines and chemokines amongst which Tumor Necrosis Factor alpha (TNF-α), which can induce LEDGF/p75 levels [43], [44]. Of note, long period or high level exposure to TNF-α makes LEDGF/p75 levels decrease again [44], which may play a role in the difference seen between LEDGF/p75 mRNA and protein levels.
Apart from the reasoning in the previous paragraph, mRNA and protein levels may differ as mRNA is not necessarily translated into (functional) protein [45], and external factors may act differently upon protein and mRNA levels [39], [46]. Our data e.g. also show a difference in mRNA versus protein expression for LEDGF/p75 in HESN (Figure 1A). Nevertheless, we did observe some correlations between protein levels in (CD4+) lymphocytes, and mRNA levels in PBMC which are predominantly composed of lymphocytes (Figure 1B–E). Of note, due to the limited number of CD4+ cells in HIV-1-infected subjects, we were obliged to work with PBMC for quantification of mRNA levels. On the other hand, the flow cytometry based method allowed us to investigate protein levels in PBMC subsets. As earlier studies also focused on mRNA levels in PBMC, we could easily compare our results with these results previously obtained. For example, our data supported that APOBEC3G mRNA levels are similar for HESN and HC [19]. On the other hand, in literature, limited information is forehand regarding protein expression in particular cell-types while proteins are considered to be of greater relevance in the “direct” fight against HIV-1 infection as they are the “work horses” of the cell.
In summary, we observed significantly lower levels of LEDGF/p75 in CD4+ lymphocytes of HESN subjects, suggesting that LEDGF/p75 may play a role in the in vivo resistance to HIV-1 infection. Untreated HIV-1-infected patients showed increased levels of the antiviral proteins APOBEC3G, TRIM5α, and tetherin, with tetherin in particular as a possible marker of advanced HIV disease. Future studies of LEDGF/p75 expression in other populations of HESN subject will be required to confirm our findings. Understanding the role of HIV-1-related host factors in HIV-1 susceptibility and disease could be of great interest for the development of future antiviral therapies.
Acknowledgments
We thank the individuals who kindly participated in the study; Dr Ndeye Fatou Ngom Gueye, Dr Ibrahima Ndiaye, and Dr Khady Ba Fall for recruiting and Marianne Ndiaye for counseling and follow-up of the Senegalese participants; Dr Aïssatou Gaye Ndiaye for HIV serological testing; and Katrien Fransen for HIV-1 viral load and HSV-2 testing. We acknowledge Chugai Pharmaceutical Co. (Japan) for kindly providing the anti-tetherin antibody and the NIH AIDS Research and Reference Reagent Program (Division of AIDS, NIAID, NIH) for providing the anti-APOBEC3G and anti-TRIM5α antibodies.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work is supported by the fund for Scientific Research (FWO) Flanders (www.fwo.be) (grant G.0660.06) and an FWO doctoral fellowship to KM. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bushman FD, Malani N, Fernandes J, D'Orso I, Cagney G, et al. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 2009;5:e1000437. doi: 10.1371/journal.ppat.1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MacPherson JI, Dickerson JE, Pinney JW, Robertson DL. Patterns of HIV-1 protein interaction identify perturbed host-cellular subsystems. PLoS Comput Biol. 2010;6:e1000863. doi: 10.1371/journal.pcbi.1000863. 10.1371/journal.pcbi.1000863 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strebel K, Luban J, Jeang KT. Human cellular restriction factors that target HIV-1 replication. BMC Med. 2009;7:48. doi: 10.1186/1741-7015-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherepanov P, Maertens G, Proost P, Devreese B, Van BJ, et al. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J Biol Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 5.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 6.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 7.Neil SJ, Zang T, Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. nature06553 [pii];10.1038/nature06553 [doi] [DOI] [PubMed] [Google Scholar]
- 8.Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, et al. A role for LEDGF/p75 in targeting HIV DNA integration. Nat Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 9.Van Maele B, Busschots K, Vandekerckhove L, Christ F, Debyser Z. Cellular co-factors of HIV-1 integration. Trends Biochem Sci. 2006;31:98–105. doi: 10.1016/j.tibs.2005.12.002. S0968-0004(05)00346-4 [pii];10.1016/j.tibs.2005.12.002 [doi] [DOI] [PubMed] [Google Scholar]
- 10.Zhang X, Kondo M, Chen J, Miyoshi H, Suzuki H, et al. Inhibitory effect of human TRIM5alpha on HIV-1 production. Microbes Infect. 2010;12:768–777. doi: 10.1016/j.micinf.2010.05.004. S1286-4579(10)00131-0 [pii];10.1016/j.micinf.2010.05.004 [doi] [DOI] [PubMed] [Google Scholar]
- 11.Black LR, Aiken C. TRIM5alpha disrupts the structure of assembled HIV-1 capsid complexes in vitro. J Virol. 2010;84:6564–6569. doi: 10.1128/JVI.00210-10. JVI.00210-10 [pii];10.1128/JVI.00210-10 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stremlau M, Perron M, Lee M, Li Y, Song B, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci U S A. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pertel T, Hausmann S, Morger D, Zuger S, Guerra J, et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature. 2011;472:361–365. doi: 10.1038/nature09976. nature09976 [pii];10.1038/nature09976 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin X, Brooks A, Chen H, Bennett R, Reichman R, et al. APOBEC3G/CEM15 (hA3G) mRNA levels associate inversely with human immunodeficiency virus viremia. J Virol. 2005;79:11513–11516. doi: 10.1128/JVI.79.17.11513-11516.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ulenga NK, Sarr AD, Thakore-Meloni S, Sankale JL, Eisen G, et al. Relationship between human immunodeficiency type 1 infection and expression of human APOBEC3G and APOBEC3F. J Infect Dis. 2008;198:486–492. doi: 10.1086/590212. 10.1086/590212 [doi] [DOI] [PubMed] [Google Scholar]
- 16.Vazquez-Perez JA, Ormsby CE, Hernandez-Juan R, Torres KJ, Reyes-Teran G. APOBEC3G mRNA expression in exposed seronegative and early stage HIV infected individuals decreases with removal of exposure and with disease progression. Retrovirology. 2009;6:23. doi: 10.1186/1742-4690-6-23. 1742-4690-6-23 [pii];10.1186/1742-4690-6-23 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho SJ, Drechsler H, Burke RC, Arens MQ, Powderly W, et al. APOBEC3F and APOBEC3G mRNA levels do not correlate with human immunodeficiency virus type 1 plasma viremia or CD4+ T-cell count. J Virol. 2006;80:2069–2072. doi: 10.1128/JVI.80.4.2069-2072.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gandhi SK, Siliciano JD, Bailey JR, Siliciano RF, Blankson JN. Role of APOBEC3G/F-mediated hypermutation in the control of human immunodeficiency virus type 1 in elite suppressors. J Virol. 2008;82:3125–3130. doi: 10.1128/JVI.01533-07. JVI.01533-07 [pii];10.1128/JVI.01533-07 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reddy K, Winkler CA, Werner L, Mlisana K, Abdool Karim SS, et al. APOBEC3G expression is dysregulated in primary HIV-1 infection and polymorphic variants influence CD4+ T-cell counts and plasma viral load. AIDS. 2010;24:195–204. doi: 10.1097/QAD.0b013e3283353bba. 10.1097/QAD.0b013e3283353bba [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sewram S, Singh R, Kormuth E, Werner L, Mlisana K, et al. Human TRIM5alpha expression levels and reduced susceptibility to HIV-1 infection. J Infect Dis. 2009;199:1657–1663. doi: 10.1086/598861. 10.1086/598861 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biasin M, Piacentini L, Caputo SL, Kanari Y, Magri G, et al. Apolipoprotein B mRNA-Editing Enzyme, Catalytic Polypeptide-Like 3G: A Possible Role in the Resistance to HIV of HIV-Exposed Seronegative Individuals. J Infect Dis. 2007;195:960–964. doi: 10.1086/511988. [DOI] [PubMed] [Google Scholar]
- 22.Miyazawa M, Lopalco L, Mazzotta F, Lo CS, Veas F, et al. The ‘immunologic advantage’ of HIV-exposed seronegative individuals. AIDS. 2009;23:161–175. doi: 10.1097/QAD.0b013e3283196a80. 10.1097/QAD.0b013e3283196a80 [doi];00002030-200901140-00002 [pii] [DOI] [PubMed] [Google Scholar]
- 23.Camara M, Dieye TN, Seydi M, Diallo AA, Fall M, et al. Low-level CD4+ T cell activation in HIV-exposed seronegative subjects: influence of gender and condom use. J Infect Dis. 2010;201:835–842. doi: 10.1086/651000. 10.1086/651000 [doi] [DOI] [PubMed] [Google Scholar]
- 24.Jennes W, Camara M, Dieye T, Mboup S, Kestens L. Higher homologous and lower cross-reactive Gag-specific T-cell responses in human immunodeficiency virus type 2 (HIV-2) than in HIV-1 infection. J Virol. 2008;82:8619–8628. doi: 10.1128/JVI.00027-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mous K, Jennes W, De RA, Pintelon I, Kestens L, et al. Intracellular detection of differential APOBEC3G, TRIM5alpha, and LEDGF/p75 protein expression in peripheral blood by flow cytometry. J Immunol Methods. 2011 doi: 10.1016/j.jim.2011.06.028. S0022-1759(11)00152-9 [pii];10.1016/j.jim.2011.06.028 [doi] [DOI] [PubMed] [Google Scholar]
- 26.McLaren PJ, Ball TB, Wachihi C, Jaoko W, Kelvin DJ, et al. HIV-exposed seronegative commercial sex workers show a quiescent phenotype in the CD4+ T cell compartment and reduced expression of HIV-dependent host factors. J Infect Dis. 2010;202(Suppl 3):S339–S344. doi: 10.1086/655968. 10.1086/655968 [doi] [DOI] [PubMed] [Google Scholar]
- 27.Madlala P, Gijsbers R, Christ F, Hombrouck A, Werner L, et al. Association of Polymorphisms in the LEDGF/p75 Gene (PSIP1) with Susceptibility to HIV-1 Infection and Disease Progression. AIDS. 2011 doi: 10.1097/QAD.0b013e328349c693. 10.1097/QAD.0b013e328349c693 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marshall HM, Ronen K, Berry C, Llano M, Sutherland H, et al. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS One. 2007;2:e1340. doi: 10.1371/journal.pone.0001340. 10.1371/journal.pone.0001340 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, et al. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. 21/14/1767 [pii];10.1101/gad.1565107 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng Y, Ao Z, Jayappa KD, Yao X. Characterization of the HIV-1 integrase chromatin- and LEDGF/p75-binding abilities by mutagenic analysis within the catalytic core domain of integrase. Virol J. 2010;7:68. doi: 10.1186/1743-422X-7-68. 1743-422X-7-68 [pii];10.1186/1743-422X-7-68 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zielske SP, Stevenson M. Modest but reproducible inhibition of human immunodeficiency virus type 1 infection in macrophages following LEDGFp75 silencing. J Virol. 2006;80:7275–7280. doi: 10.1128/JVI.02470-05. 80/14/7275 [pii];10.1128/JVI.02470-05 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coleman CM, Spearman P, Wu L. Tetherin does not significantly restrict dendritic cell-mediated HIV-1 transmission and its expression is upregulated by newly synthesized HIV-1 Nef. Retrovirology. 2011;8:26. doi: 10.1186/1742-4690-8-26. 1742-4690-8-26 [pii];10.1186/1742-4690-8-26 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jolly C, Booth NJ, Neil SJ. Cell-Cell Spread of Human Immunodeficiency Virus Type 1 Overcomes Tetherin/BST-2-Mediated Restriction in T cells. J Virol. 2010;84:12185–12199. doi: 10.1128/JVI.01447-10. JVI.01447-10 [pii];10.1128/JVI.01447-10 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gummuluru S, Kinsey CM, Emerman M. An in vitro rapid-turnover assay for human immunodeficiency virus type 1 replication selects for cell-to-cell spread of virus. J Virol. 2000;74:10882–10891. doi: 10.1128/jvi.74.23.10882-10891.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malim MH, Emerman M. HIV-1 accessory proteins–ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3:388–398. doi: 10.1016/j.chom.2008.04.008. S1931-3128(08)00126-1 [pii];10.1016/j.chom.2008.04.008 [doi] [DOI] [PubMed] [Google Scholar]
- 36.Tokarev A, Skasko M, Fitzpatrick K, Guatelli J. Antiviral activity of the interferon-induced cellular protein BST-2/tetherin. AIDS Res Hum Retroviruses. 2009;25:1197–1210. doi: 10.1089/aid.2009.0253. 10.1089/aid.2009.0253 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen K, Huang J, Zhang C, Huang S, Nunnari G, et al. Alpha interferon potently enhances the anti-human immunodeficiency virus type 1 activity of APOBEC3G in resting primary CD4 T cells. J Virol. 2006;80:7645–7657. doi: 10.1128/JVI.00206-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stopak KS, Chiu YL, Kropp J, Grant RM, Greene WC. Distinct patterns of cytokine regulation of APOBEC3G expression and activity in primary lymphocytes, macrophages, and dendritic cells. J Biol Chem. 2007;282:3539–3546. doi: 10.1074/jbc.M610138200. M610138200 [pii];10.1074/jbc.M610138200 [doi] [DOI] [PubMed] [Google Scholar]
- 39.Koning FA, Newman EN, Kim EY, Kunstman KJ, Wolinsky SM, et al. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J Virol. 2009;83:9474–9485. doi: 10.1128/JVI.01089-09. JVI.01089-09 [pii];10.1128/JVI.01089-09 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asaoka K, Ikeda K, Hishinuma T, Horie-Inoue K, Takeda S, et al. A retrovirus restriction factor TRIM5alpha is transcriptionally regulated by interferons. Biochem Biophys Res Commun. 2005;338:1950–1956. doi: 10.1016/j.bbrc.2005.10.173. S0006-291X(05)02472-1 [pii];10.1016/j.bbrc.2005.10.173 [doi] [DOI] [PubMed] [Google Scholar]
- 41.Sakuma R, Mael AA, Ikeda Y. Alpha interferon enhances TRIM5alpha-mediated antiviral activities in human and rhesus monkey cells. J Virol. 2007;81:10201–10206. doi: 10.1128/JVI.00419-07. JVI.00419-07 [pii];10.1128/JVI.00419-07 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawai S, Azuma Y, Fujii E, Furugaki K, Ozaki S, et al. Interferon-alpha enhances CD317 expression and the antitumor activity of anti-CD317 monoclonal antibody in renal cell carcinoma xenograft models. Cancer Sci. 2008;99:2461–2466. doi: 10.1111/j.1349-7006.2008.00968.x. CAS968 [pii];10.1111/j.1349-7006.2008.00968.x [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma P, Singh DP, Fatma N, Chylack LT, Jr, Shinohara T. Activation of LEDGF gene by thermal-and oxidative-stresses. Biochem Biophys Res Commun. 2000;276:1320–1324. doi: 10.1006/bbrc.2000.3606. 10.1006/bbrc.2000.3606 [doi];S0006-291X(00)93606-4 [pii] [DOI] [PubMed] [Google Scholar]
- 44.Takamura Y, Fatma N, Kubo E, Singh DP. Regulation of heavy subunit chain of gamma-glutamylcysteine synthetase by tumor necrosis factor-alpha in lens epithelial cells: role of LEDGF/p75. Am J Physiol Cell Physiol. 2006;290:C554–C566. doi: 10.1152/ajpcell.00398.2005. 290/2/C554 [pii];10.1152/ajpcell.00398.2005 [doi] [DOI] [PubMed] [Google Scholar]
- 45.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. clinchem.2008.112797 [pii];10.1373/clinchem.2008.112797 [doi] [DOI] [PubMed] [Google Scholar]
- 46.Chiu YL, Greene WC. Multifaceted antiviral actions of APOBEC3 cytidine deaminases. Trends Immunol. 2006;27:291–297. doi: 10.1016/j.it.2006.04.003. [DOI] [PubMed] [Google Scholar]