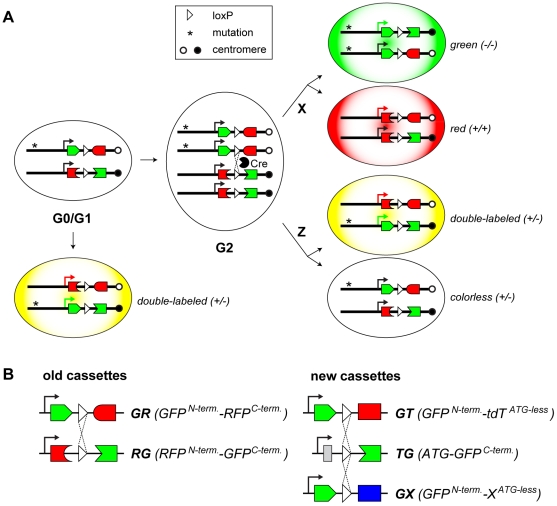
Figure 1. The MADM principle and design of new MADM cassettes.
A) MADM relies on two reciprocally chimeric marker genes (for example, GR and RG, see part B below for cassette description) that have been knocked into the same locus on homologous chromosomes. Recombination in the G2 phase of the cell cycle regenerates the functional marker genes on a pair of chromatids. X-segregation of chromatids (the recombinant chromatids segregate to different cells) generates a red and a green cell. Z-segregation of chromatids (the recombinant chromatids congregate to the same cell) generates a double-labeled (yellow) cell and an unlabeled cell. If a mutation (asterisk) is present distally to the GR cassette, the green cells will be homozygous for the mutation. This orientation of the cassettes corresponds to MADM in the Rosa26 locus. If the cassettes are in the opposite orientation with respect to the centromere, the genotypes for green and red cells will be inverted (for example in MADM-11). If mitotic recombination occurs in G0 or G1, a double-labeled cell is produced without altering the genotype of the cell. B) The “old” MADM cassettes contained two genes encoding fluorescent proteins (dsRed2 and GFP) split roughly in the middle. The “new” cassettes use the same GFP split, but split the second gene (for example, tdTomato) into ATG and GeneATG-less. That way, the ATG-GC-terminus (for simplicity, TG) becomes a universal cassette that can be paired with any G-GeneATG-less cassette. The single white triangle represents a single loxP site, a combination of loxP sites or the loxP-flanked (floxed) neomycin resistance gene (see Figure S1 for detailed description of MADM cassettes).