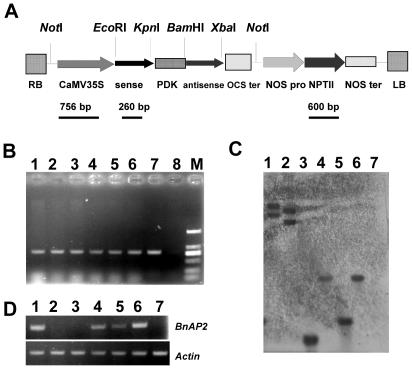
Figure 6. Transformation of B. napus cultivar Zhongshuang 6 with intron-spliced hairpin RNA (ihpRNA) as RNAi construct that targets BnAP2 gene.
A, Schematic diagram of T-DNA of the binary vector pBnAP2-RNAi. RB, right border; CaMV 35S, CaMV 35S promoter; Anti-sense, fragment in anti-sense orientation; PDK, PDK intron; sense, fragment in sense orientation; OCS ter, OCS terminator; NOS pro, NOS promoter; NPT II, neomycin phosphotransferase gene; NOS ter, NOS terminator; LB, left border. The sites of a 756-bp probe used for Southern blot, a 600-bp fragment used for PCR template and a 260-bp used for RT-PCR were indicated. B, PCR analysis of the RNAi construct transgenic plants using genomic DNA as the template. Lane 1, positive control (pBnAP2-RNAi); lane 8, untransformed Zhongshuang 6 (CK1); lanes 2–7, the tested transgenic plants BnAP2-RNAi-1, BnAP2-RNAi-2, BnAP2-RNAi-3, BnAP2-RNAi-4, BnAP2-RNAi-18 and BnAP2-RNAi-26, respectively; M, DL 2000 marker. C, Southern-blot analysis of the BnAP2-RNAi transgenic plants. Lane M, DNA marker (unit, bp); lane 7, untransformed Zhongshuang 6 (CK1); lanes 1–6, the tested transgenic plants BnAP2-RNAi-1, BnAP2-RNAi-2, BnAP2-RNAi-3, BnAP2-RNAi-4, BnAP2-RNAi-18 and BnAP2-RNAi-26, respectively. Genomic DNA from all transgenic and CK1 plants was digested by KpnI. D, RT-PCR analysis of the BnAP2 gene in the BnAP2-RNAi transgenic plants. lane 1, untransformed Zhongshuang 6 (CK1); lanes 2–7, the tested transgenic plants BnAP2-RNAi-26, BnAP2-RNAi-18, BnAP2-RNAi-4, BnAP2-RNAi-3, BnAP2-RNAi-2 and BnAP2-RNAi-1, respectively.