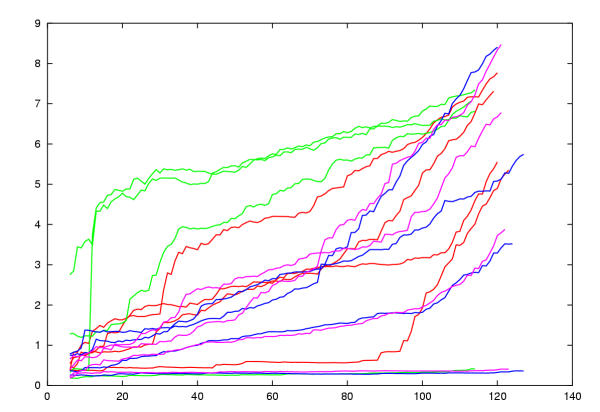
Figure 7.
Control model uRMSd values. Each model was compared with its known structure and the local residue matches calculated by SAP were ranked. The uRMSd was calculated for increasingly larger sets of ranked residues and plotted against set size. This means that the best fitted residues in each comparison are found to the left of the plot with increasing divergence towards the right. Four models are plotted for each of four CA proteins of known structure: 1d1dA = red, 1qrjA = green, 1eia = purple, 1e6jP = blue. The best models are those of sequences built on their own structure. Three of these remain low throughout their length while one has poor loops and rises towards the right. Above these are two models for the most similar sequences of 1e6jP and 1eia built on each others structure (22% identity). Most models lie in the mid range with a few accumulating early errors due to shifts in the core regions.