Abstract
Helper T cell differentiation occurs in the context of the extracelluar cytokine milieu evoked by diverse microbes and other pathogenic stimuli along with T cell receptor stimulation. The culmination of these signals results in specification of T helper lineages, which occurs through the combinatorial action of multiple transcription factors that establish distinctive transcriptomes. In this manner, inducible, but constitutively active master regulators work in conjunction with factors like the signal transducer and activator of transcriptions (STATs) that sense the extracellular environment. The acquisition of distinctive transcriptome is also dependent upon chromatin modifications that impact key cis elements as well as the changes in global genomic organization. Thus, signal transduction and epigenetics are linked in these processes of differentiation. In this review, recent advances in understanding T helper lineage specification and deciphering the action of transcription factors are summarized with emphasis on comprehensive views of the dynamic T cell epigenome.
Introduction
CD4+ T cells can differentiate into an array of effector, helper and regulatory T cells, and the range of possibilities for a CD4+ T cell seems to keep expanding(1, 2). These differentiation processes are critical for host defense and immunoregulation, but also represent a remarkably simple and tractable model system for understanding basic principals in cellular specification and gene regulation. While CD4+ T cell subsets have elements of stability and have been referred to as distinct lineages, there are increasing examples of flexibility among them. This raises fundamental questions as to what factors control stability and permit plasticity in cellular phenotype.
For all cells, the process of differentiation represents the integration of intrinsic and extrinsic factors that control cell fate commitment. In the case of differentiating CD4+ T cells, much has been learned about the trans acting factors that drive lineage commitment. The major players will be discussed briefly, but we refer readers to the many excellent reviews on this subject (1-3). In addition, much effort over a number of years has given us a great deal of information on the cis elements that control the expression of key lineage-defining genes, principally the Il4 (3) and Ifng (1) genes. Again, the interested reader is referred to outstanding, detailed reviews on structure of these key immunologic genes and we apologize in advance that we are unable to cite the many seminal studies that have led to our present understanding.
In this review our focus will be on epigenetics and helper T cell differentiation. However, this is also not a new topic; as will be discussed, the concept of epigenetics comes from the mid 20th century.and there are already several well-written reviews on this topic as it relates to T cells (1, 2). What is rapidly changing is our biochemical understanding of epigenetic process and our ability to map global, genomewide changes in the epigenome. In other words, we have begun to characterize the epigenome of differentiating helper T cells. Since this topic was last reviewed in Annual Reviews of Immunology (2, 4), many technological advances have become commonplace that permit comprehensive views of the “epigenetic landscape” rather than snapshot views of portions of single genes. Consequently, we will focus on the technologies that provide these new ways of looking at helper T cells along with the evolving information that has been provided.
What elements define T helper lineages?
Naïve CD4+ T cells differentiate into distinct T helper lineages, whose phenotypes are commonly defined by the signature effector cytokines produced, the master transcription factors expressed and the type of microbial pathogens controlled. Specifically, T helper 1 cells (Th1) express interferon-γ (IFN-γ) and the transcription factor T-bet and serve to control intracellular pathogens. Th2 cells express interleukin-4 (IL-4) and Gata3 and limit helminth infestations. Th17 cells produce IL-17, express Rorc and protect against extracellular bacteria and fungi.
In addition to selective expression of cytokines, another critical aspect of the biology of differentiating helper T cells is the silencing of lineage-inappropriate cytokines. Consequently, as Th1 are fully polarized to become efficient IFN-γ producers, their ability to produce IL-4 and IL-17 is effectively repressed. Beyond their ability to become effector cells, CD4+ T cells can become FoxP3-expressing regulatory T cells (Treg) that regulate immune homeostasis.
Less well defined than the class Th subsets are follicular helper cells (Tfh). This subset of cells localize to B cell follicles and are critical for providing B cell help, meaning they have essential functions in promoting class switching of B cells. Their signature cytokine is IL-21 and they express the transcriptional repressor, Bcl-6. However, IL-21 and Bcl-6 are not uniquely expressed by Tfh cells and the relationship between Tfh and other Th subsets is still an area of intense investigation (5). Other T cell subsets include Th22 and Th9 cells, which preferentially express IL-22 and IL-9 (6, 7). It appears that the transcription factor Pu.1 is important for Th9 cells (8). The extent to which we should be viewing these subsets as lineages, per se, remains debatable.
While this simple scenario (expression of lineage defining master regulator and signature cytokine) has proven useful, it has become increasingly clear that CD4+ T cells can express more than one master regulator. Illustrative examples include Foxp3+/T-bet+ and Rorc+/T-bet cells (9). Historically, the definition of what it is to be a Th1 or Th2 is also a bit tautologic argument: A Th1 cell makes IFN-γ. How do we know it’s a Th1 cell? It makes IFN-γ. The work of Hegazy and colleagues make the problem of such a definition clear. They showed that committed Gata-3+Th2 cells that are adoptively transferred into virally-infected mice are reprogrammed to adopt Gata-3+T-bet+ and IL-4+IFN-γ+ phenotypes (10). So what do we call a Gata-3+T-bet+ and IL-4+IFN-γ+ cell? In the present nosology there is no simple solution and the present reality is that CD4+ T cells exhibit rather substantial plasticity. In fact, this may be advantageous in terms of host defense.
In the same vein, it also not a simple task to understand exactly what the extended phenotype of Th cell is. In some cases, expression of particular chemokines and chemokine receptors are associated with different Th subsets; however, there are also good examples of such factors being more promiscuously expressed. As discussed in more detail below, there is not a simple correlation between cytokine, master regulator and a precise cassette of other lineage-defining genes. In this regard, it will be quite informative to more carefully define T helper lineage not only by a small number of cytokine/transcription factor expression, but also with more comprehensive transcriptome signatures. There is no shortage of microarray studies of different Th subsets; however, the transcriptomes obtained offer glimpses at the global state of RNA expression at very limited times. Highly dynamic global gene expression will require many of transcriptome snapshots to permit a complete understanding of kinetics of transition to different states of differentiation.
Transcription factors as drivers of T helper differentiation
In general terms, cell phenotype is often the result of expression of master regulator transcription factors. In non-T cells, vivid examples include master regulators such as MyoD or Pax family transcription factors (11). Expression of such transcription factors is necessary and sufficient for cell fate determination and those factors bind directly to target genes involved in specification.
The differentiation of naïve T cells into specific helper lineages require cues from extracelluar environment and this comes in the form of antigen, co-stimulatory molecules, adhesion molecules as well as various cytokines. Extrinsic instructions received by naive CD4+ T cells during initial engagement with antigen-presenting cells are then converted into cell-intrinsic changes. There are several combinations of transcription factors that contribute to establish each T helper subset.
Factors that promote Th1 differentiation
For Th1 cells, Stat1 and Stat4 are both important factors, receiving signals from interferons (IFN) and IL-12 respectively. Stat1 and Stat4 can both induce T-bet, which works in conjuction with Hlx, Runx3, and Ets family members to promote IFN-γ production and repress IL-4 transcription. T-bet also inhibits the function of Gata3 and Rorc, thereby antagonizing Th2 and Th17 differentiation. T-bet is phosphorylated by Itk, and phosphorylated T-bet binds and repress Gata3, thereby interfering with the Gata3’s function (12). It should be noted, however, that Th1 differentiation can occur in the absence of the cognate master regulator T-bet or Stat4 through other pathways. In CD8 cells, a different T-box family member, Eomes, is the major regulator of IFN-γ production.
Factors that drive Th2 differentiation
For Th2 cells, Stat5 or Stat6 can initiate specification by induction of Gata3, but Notch signaling can also induce Gata3 in a Stat6-independent manner (13). GATA3 in turn induces c-Maf, which promotes Th2 differentiation; however, c-Maf deficiency does not abrogate production of Th2 cytokines other than IL-4, suggesting a selective role of c-Maf in regulating IL-4 transcription (14). Additionally, STAT3 has been found to be a contributor to Th2 differentiation (15).
Factors important for Th17 differentiation
Cytokines like IL-6 and IL-23 activate Stat3, which induce Rorc, the master regulator for Th17 cells. In addition, other transcription factors including aryl hydrocarbon receptor, Batf, IkappaBzeta, IRF-4 and Runx1 are reported to promote IL-17 expression. In contract, Foxp3, Gfi-1 and Ets-1 are reported to inhibit Th17 cell differentiation. Tcf-1 is another repressor of Th17 that acts by directly binding and repressing the Il17 gene. Tcf-1 also negatively regulates IL-7R on Th17 cells and constrains the survival of inflammatory Th17 cells (16).
Factors involved in Treg cell differentiation
IL-2 plays a pivotal role to maintain Treg population. Activated by IL-2, Stat5 binds the Foxp3 gene and induces its expression and Foxp3, in turn, directly regulates many genes involved in the program of Treg cells. FoxP3 can bind Rorγt and interfere with Th17-cell development (17). Runx1 affects the balance between Th17 and Treg through its ability to interact with Rorc and Foxp3. Rorc and Runx cooperate to induce its expression of IL-17, whereas Foxp3-Runx1 interactions inhibit Th17 differentiation (18). Runx proteins can also regulate the expression of Rorc (18) and FoxP3 (19). Nrfa2 is yet another factor that positively regulates FoxP3 expression (20). Foxo family members, Foxo1 and Foxo3, also participate in specifying Treg developmental pathway (21). Treg cell specific deletion of a transcription factor critical for other effector lineage, namely IRF4 for Th2 and Stat3 for Th17 cells, suggests that Treg cells elicit effector response-specific suppression by using components of the transcriptional machinery important for the “target” effector lineage (22, 23). Of note, in response to IFN-γ, some Treg cells can express T-bet which promotes expression of CXCR3 and promotes trafficking of Treg cells to sites of Th1-associated inflammation (24). Recently, it was reported that one key role of FoxP3 is to repress the genome organizer protein, SATB1. This constrains chromatin remodeling at cytokine loci for other effector lineages and thereby maintains functionality of Treg (25).
Common factors that regulate cytokine production
In addition to promotion of lineage commitment, transcription factors influence helper T cell function by directly affecting the acute production of cytokines. After differentiation to a particular subset, effectors become activated by secondary engagement of the TCR and co-stimulatory molecules, with or without cytokines. Transcription factors such as NF-κB family members, NFAT proteins, AP-1, and STAT proteins can drive acute induction of cytokine transcription following stimulation.
What elements define the epigenome?
Gene expression is not simply dependent upon presence or absence, and/or the state of activation of a transcription factor, and other factors determine whether a genomic segment can be subject to regulation by transactivating factors. For many years, even before we knew many of the details of physical nature of genes, it was appreciated that phenotype of progeny cells is preserved through multiple cell divisions and preservation of phenotype occurs over time and in various environments. Despite the vast differences in phenotype of all the various cells in tissues and organs, the underlying DNA sequence remains the same. So what explains the differences and what preserves stability of phenotype?
Historical views on epigenome
Waddington in the mid 20th century first used the term “epigenetics” as a manifestation of genetic activity (26). At this time though, Waddington did not have an appreciation of what a gene really was. With time the term epigenetics has come to describe heritable changes in phenotype or gene expression without changes in DNA sequences itself. It has been recognized that terminally differentiated cells can autonomously maintain their distinctive features through mitotic divisions even in the absence of exogenous signals or the action of transcription factors that initially induced the phenotype. Thus the term epigenetics refers to underlying mechanisms that preserve cellular memory and maintain distinctive transcriptional profiles and cellular identity. More recently the term epigenetics has come to have a more biochemical meaning. Some of the elements that constitute epigenetic information that include: (1) histone tail modifications and histone variants, (2) DNA methylation, (3) nucleosome compaction and (4) chromatin interaction/chromosome conformation. Broadly speaking the term can also refer to other mechanisms such as the action of long non-coding RNAs and microRNAs. We will discuss each of these topics individually.
Histone tail modification
Posttranslational modifications of histones, particularly at N-terminal tails, present myriad patterns of epigenetic information through enzyme-mediated acetylation, methylation, phosphorylation, ubiquitination, sumoylation etc. Enzymes that add or remove those modifications have also been identified and in some cases, targeted genetic perturbation of those enzymes result in alterations of phenotype related to T cell function (Table 1). While numerous differential combinations of histone marks could potentially exist, it has been noted that a few select combinations of marks are quite useful and powerful enough to profile functionality of histone epigenome. Such representative marks include: H3K4 mono/di/trimethylation associated with permissive active conformation, H3K27 trimethylation associated with repressive conformation, and H3K36 trimethylation associated with actively transcribed coding regions. In particular, two histone methylation marks at H3K4 and H3K27 represent opposite functions, active chromatin versus silenced chromatin, and they are associated with trithorax (27) and polycomb (28) complexes, respectively, that are responsible to “write” those marks through the catalytic subunit, methytransferases MLL and Ezh2, respectively. There are also demethylases (LSD1 and Jumonji C domain family proteins) that erase those marks (29). As will be discussed, genome-wide mapping of those key histone marks and subsequent combinatorial analyses can distinguish the status of chromatin as 3 different stages: silent, poised, and active (1). “Poised” status represents an intermediate state between active and silent, and is defined by either null histone mark or by simultaneous presence of both active and repressive marks called as bivalent. Similar combinatorial analysis of histone marks can be successfully used to predict regions that actively transcribing non-coding RNAs (30). Histone acetylation is also important for chromatin function and gene transcription, and two classes of antagonizing enzymes contribute to creating and removing these marks, namely histone acetyltransferases (HATs) and deacetylases (HDACs), respectively (31). The classic view relates HAT activity and acetylated chromatin to active transcription and HDAC activity with repressed transcription. However, the genome-wide distribution of HATs and HDACs provided a more complex picture pointing to an involvement of both HATs and HDACs with active transcription (32). Thus dynamic interplay between HATs and HDACs on the same loci seems to be a key for maintaining the state of histone acetylation and active transcription (31). As canonical histone (H2A, H2B, H3 and H4) are packed into nucleosomes to form building blocks for chromatin, variant histones can perform additional specialized functions in transcriptional regulation and beyond including DNA repair, chromosome segregation and others (33).
Table 1.
Immunological phenotypes for mice deficient in various chromatin modifying/remodeling proteins in T cells.
| Protein/Inhibit or |
Species | details | phenotype | Reference |
|---|---|---|---|---|
| BRG1 | Murine (knock out mice) |
the ATPase subunit of the SWI/SNF complex |
Abnormal T cell development and IFN-γ production |
Gebuhr (JEM, 2003), Chi (Immunity 2003) |
| MLL | Murine (heterozygous mice) |
Trithorax component (H3K4 methyltransferase) |
Defect in maintenance of Gata3, Il4, Il5, and Il13 | Yamashita (Immunity, 2006) |
| MENIN | Murine (knock out mice) |
Trithorax component | Impaired the maintenance of Gata3 expression | Onodera (JEM, 2010) |
| BMI1 | Murine (knock out mice) |
PRC1 component | Enhanced expression of Noxa gene and reduced memory T cell subsets |
Yamashita (JEM, 2008) |
| MEL18 | Murine (knock out mice) |
PRC1 component | Impaired Gata3 induction | Kimura (Immunity, 2001) |
| EZH2 | Murine (knock out mice) |
PRC2 component | Defect in actin polymerization-dependent processes such as antigen receptor signaling in T cells |
Su (Cell, 2005) |
| BPTF | Murine (knock out mice) |
NURF component | Defects in thymocyte development | Landry (Gene and Dev. , 2011) |
| BAF57 | Murine (dominant negative mutation transgenic) |
BAF component | Impaired T cell development | Chi (Nature 2002) |
| DNMT1 | Murine (knock out mice) |
DNA methyltransferases | Increased Th1 and Th2 cytokine production | Makar (NI, 2003), Lee (Immunity, 2001) |
| MBD2 | Murine (knock out mice) |
Methyl-DNA binding protein |
Ectopic IL-4 expression | Hutchins (Mol Cell, 2002) |
DNA methylation
DNA methylation on cytosines in CpG dinucletides is another form of epigenetic information that can repress transcription when introduced at gene promoters. Through the action of DNMT1, the pattern of DNA methylation is faithfully copied from one generation to the next and is thus considered to be the most stable epigenetic mark that confers heritable epigenetic memory. Global distributions of DNA methylation, collectively called as DNA methylomes, for several organisms have now been reported. Genome-wide distribution of DNA methylation pattern revealed methylation on CG as well as non-CG context, and the former mainly was associated with promoter region and the latter was with gene body region that was actively transcribed in ES cells (34). Recent comparative profiling of DNA methylomes between embryonic stem (ES) cells and induced pluripotent stem cells (iPS) revealed clear epigenetic difference between the cells and raised a question regarding the reprogramming of iPS cells. The discrepancy in profiles of methylomes suggested incomplete reprogramming of iPS cells and incomplete erasure of previous phenotypic memory of the original cell type (35). While the identities and function of DNA methyltransferases are well characterized (36), the mechanism to achieve DNA demethylation remains to be clarified as no single enzyme or mechanism has gained decisive biochemical and genetic support, and it is plausible that multiple mechanisms exist to carry out the task depending on specific biological context (37). Recently 5-hydroxymethylation of cytosine has been reported as a new epigenetic mechanism crucial for regulating pluripotency and differentiation of ES cells and hematopoietic stem cells. The genome-wide distribution of 5-hydroxymethylcytosine (5hmC) showed deposition of the mark in transcribed regions, transcription start sites and a subset of enhancer regions (38, 39). TET (ten eleven translocation) family proteins are enzymatically responsible for converting 5-methylcytosine (5mC) to 5hmC but can also convert 5mC to 5-formyl-C or 5-carboxy-C. This raises the possibility that TET proteins contribute to DNA demethylation reactions (40).
Nucleosome compaction
Most genomic DNA in eukaryotes is incorporated into nucleosomes and folded in a tightly compacted fashion(41). Nuclesome positioning, referring to the localization of an individual histone octamer with respect to a specific DNA sequence, modulates accessibility of regulatory proteins to DNA and influences eukaryotic gene regulation, thus constitutes epigenetic information (42). Most transcribed regions have reduced nucleosome occupancy over the promoter, and these nucleosome-depleted regions (NDRs) are generally flanked by two well-positioned nucleosomes (43).
ATP-dependent remodeling has been considered as important modulators of chromatin structure and nucleosome dynamics (44, 45). Various enzymatic machines can perturb intrinsic histone-DNA interactions. These nucleosome remodelers are multi-protein complexes that employ ATP hydrolysis to slide or disassemble histone octamers (43). There are several classes of chromatin remodelers, including the SWI/SNF (BAF), ISWI, INO80, SWR, and Mi-2/CHD, each specialized for a different function that can positively or negatively regulate transcription (46, 47). The active ATPase motors of these large complexes associate with a variety of other proteins that regulate their activity and help target them to chromatin.
Chromatin interaction and chromosome conformation
Chromatin looping generated by chromosome interaction from distance has been recognized as another mechanism to affect regional transcription (48). Chromatin looping brings multiple distal regulatory elements such as enhancers into close contact with the promoter of the genes that they regulate. Such interactions can occur in cis in a linear fashion on the same chromosome, as reported for the Th2 cytokine locus and Th2 locus control region (LCR) in T cells (49). However, interactions can also occurs in trans, meaning that regions of different chromosomes can interact. For instance the Ifng locus on chromosome 10 and Il4 locus on chromosome 11 can associate in naïve T cells (50). This results in the formation of three-dimensional structures that promote efficient enrichment of functional components for transcription including transcription activators, RNA polymerase II, component of pre-initiation complexes, etc. Such functional units have been termed transcription factories (51) and potentially provide a mechanism for co-regulation of diverse genes on different chromosomes.
Cis Regulatory Elements and Epigenetic Modifications
Cis regulatory elements are the DNA elements that control gene expression and constitute an integral part of gene structure. Cis regulatory elements usually contain the binding site for trans-acting factors and serve as the site of epigenetic modification as well. Functionally relevant cis-elements may be located 5′ to the coding sequence of the gene it controls (in the promoter region or further upstream), in an intron, or 3′ to the gene’s coding sequences. In differentiating Th cells, the focus has been on the structure of lineage-defining cytokine genes including their distal cis regulatory elements.
Structure of the Ifng gene
The nearest gene neighbor of the Ifng and IFNG genes in mouse and human is approximately 420 and 500 kb downstream. Upstream, the nearest functional gene is IL26 in human and Il22 in mouse (due to near complete deletion of IL26 in rodents), which resides approximately 245 kb upstream of Ifng. IL-22 and IL-26 are IL-10 family cytokines, which are not expressed in Th1 cells. This implies the existence of an insulator or boundary element between these genes and the Ifng/IFNG loci. The importance of distant cis regulatory elements surrounding Ifng/INFG genes was established by the creation of transgenic mice carrying a human IFNG BAC transgene comprising 90 kb of upstream and downstream flanking sequence. While mice carrying extended IFNG region recapitulated normal, regulated IFN-γ expression, including Th0/Th1/Th2 selectivity, the transgenic mice bearing a construct comprising the Ifng promoter region alone failed to reproduce fidelity in IFN-γ expression (52). Taken together, the data suggest that the complete IFNG and Ifng loci appear to encompass 180 and 140 kb in human and mouse, respectively (52, 53).
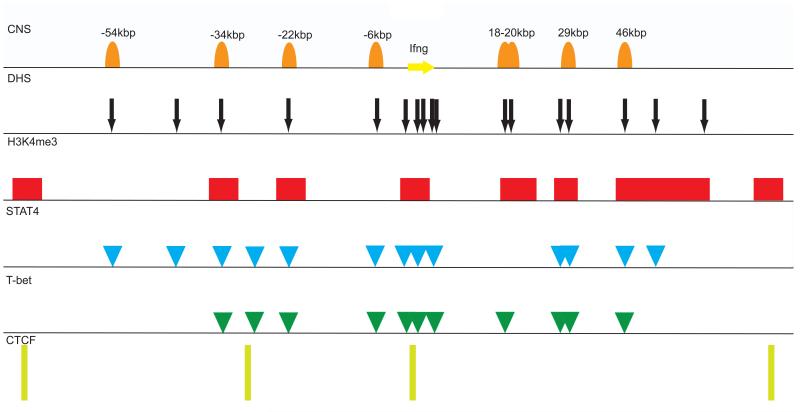
Mapping of individual cis elements in the Ifng and IFNG loci began about 20 years ago through identification of conserved non-coding sequences (CNS) combined with DNAse hypersensitivity assays. Further information on histone epigenetic marks, DNA methylation, transcription factor binding or chromosome conformation have added accuracy and functionality to each elements among which CNS-34, CNS-22, CNS-6, CNS+18-20, CNS-29 have been found to be key regulatory hubs that recruit multiple trans acting factors to facilitate Th1-specific remodeling of the Ifng locus (54-56)(see also Figure 1). Comprehensive reviews on the regulatory elements of Ifng/IFNG genes in human and mice are found in (1, 57).
Fig. 1.
The mouse Ifng locus and surrounding regulatory regions identified by various chromatin signatures in Th1 cells. CNS denotes the conserved noncoding sequence described previously (1). DNase hypersensitivity sites (DHS) and T-bet binding sites are depicted from genome-wide data available from Gene Expression Omnibus (GEO) under the accession number GSE33802. H3K4me3 denotes trimethylation of lysine 4 on histone 3 that is regarded as permissive histone modification. Locations of H4K4me3 and STAT4 binding sites were depicted from genome-wide data in GEO with the accession number GSE22105. Location of CCCTC-binding factor (CTCF) binding sites was described (1) and also confirmed by genome-wide data (J. J. O., unpublished observation).
Structure of the IL4 gene
The Il4/IL4 loci appear to be equally complex, encompassing approximately 200 kb and comprising the Il4, Il13 and RAD50 genes. Downstream of RAD50 is the Il5 gene. Transgenic expression of a 30 kb construct encompassing the Il4 and Il3 genes did not exhibit appropriate regulated expression, whereas a construct containing ~25 kb downstream of RAD50 revealed the presence of a locus control region (LCR)(58). Like the Ifng locus, much work has been done to identify important cis elements within and beyond the LCR. Activation-induced DNA demethylation at the LCR has also been reported (59). Interestingly, functional deletion analysis also identified a silencer element (HS-IV), deletion of which derepressed IL-4 expression (3) and subsequently involvement of T-bet and Runx for the silencer function was revealed (60, 61). Many of the identified elements (HS-I, II, VA, V), along with each of the Th2-cytokine promoters are direct targets of NFAT, other TCR-induced transcription factors and Th2 promoting transcription factors such as STAT6 and Gata3. An extensive review of the regulatory elements in Th2 cytokine loci can be found in (4).
Cis elements involved in FoxP3 gene regulation
Treg cells do not have a signature cytokine; on the contrary, production of most cytokines is repressed. However, they do have the master regulatory transcription factor FoxP3. A conserved CpG rich region denoted as a Treg-specific demethylated region (TSDR) resides upstream of the FoxP3 exon1 (62) and also another CpG rich region was found in the 1st intron(63). This TSDR is demethylated in nTreg cells and is associated with stable expression of FoxP3 (62, 64). In contrast, there is residual methylation of this region in iTreg cells, consistent with instability of FoxP3 expression (64). Further fine mapping with in vivo functional deletion analysis identified 3 major cis elements, CNS-1, CNS-2 (both in upsteam of exon 1) and CNS-3 (in intron 1) with discrete functions (65). CNS-1 that contains a TGFβ-NFAT response element is important for iTreg generation in gut. CNS2 recruits FoxP3-Runx/CBFβ complex in a CpG demethylation-dependent manner and serves as a cellular memory module for Treg lineage stability. CNS3 is a pioneer element that binds c-Rel homodimer activated downstream of TCR/CD28 and serves to open up the FoxP3 locus early in development and therefore is indispensable for normal thymic and peripheral Treg generation.
Epigenetics and T cell differentiation – cause or effect?
The first evidence to support epigenetic contribution in gene regulation came with drugs that affected the status of epigenetic information. The DNA methylation inhibitor, 5-azacytidine , caused constitutive production of IL-2 (66) and IFN-γ (67) and HDAC inhibitors enhanced production of Th1 and Th2 cytokines (68-70). These data suggested that modulation of epigenetic information on DNA or histones could lead to change in cytokine production.
Genetic approaches by deletion of chromatin modifying/remodeling proteins have added more evidence for the causal roles of epigenetic modifications in regulating cytokine expression (Table 1). Conditional ablation of DNMT1 (DNA methyltransferase)(71, 72) or MbD2 (methyl-CpG binding domain protein 2)(73) led to increased expression of IFN-γ and Th2-type cytokines and inability of Th1 or Th2 cells to properly silence the expression of opposing cytokine genes. The importance of cell cycle progression in controlling IFN-γ and IL4 expression was also noted, suggesting that for removal or establishment of local epigenetic status, proliferative signals are required in addition to differentiation signals provided by cytokines (68).
Targeting histone modifying enzymes also led to impairment of T cell development and function. Haplo-insufficiency of the H3K4 methyltransferase MLL (MLL+/−), the catalytic subunit of Trithorax group (TrxG), resulted in defective maintenance but not initial induction of Il4, Il5, Il13 and Gata3 expression in Th2 cell, indicating its crucial role for memory Th2 cell maintenance (74). Similarly, deletion of menin, another component of TrxG, showed impairment in maintaining Gata3 expression (75). Also, mice that lack MEL18, a polycomb repressor complex 1 (PRC1) protein that binds to trimethylated H3K27, have impaired GATA3 induction and Th2-cell differentiation (76), whereas deletion of Bmi1, another component of PRC1, impaired memory Th2 cell survival(77). EZH2, a major H3K27 methyltransferase, binds to and suppresses the Il4/Il13 locus in Th1 cells (78). In addition though, EZH2 has an extra-nuclear function in that it associates with cytosolic protein complex and controls T cell receptor-induced actin polymerization (79).
ATP-dependent chromatin remodeling complexes are crucial for both the assembly and disassembly of chromatin structure (45). Brahma-related gene 1 (BRG1) is a catalytic subunit of SWI/SNF remodeling complexes and T cell specific deficiency of Brg1 in mice resulted in both thymic and peripheral abnormalities of T cells (80, 81). Brg1 binds the Ifng promoter in Th1 cells and knockdown of Brg1 results in decreased IFN-γ production (82). Targeted mutation of BAF57 (a component of SWI/SNF complex) resulted in dysregulation of CD4+ gene silencing in T cells (47). Similarly T cell specific deletion of BPTF (a component of nucleosome remodeling factor NURF) resulted in developmental arrest of thymocytes (83). Overall, mounting evidence strongly argues for a causal role of epigenetic mechanisms in supporting T lineage differentiation and maintaining specification.
How is global epigenetic status evaluated?
Conventional biochemical analyses of epigenetic modifications include chromatin immunoprecipitation (ChIP), DNAse hypersensitivity (HS) assay, or bisulfite conversion of cytosines. However, these approaches have undergone a renaissance when coupled with next generation sequencing technology. Several key techniques that can address an aspect of global epigenetic information are summarized in Table 2 that include: (1) ChIP-seq, (2) MNase-seq, (3) DNase-seq, (4) Bisulfite-seq, (5) 4C/5C/Hi-C-seq, and (6) ChIA-PET-seq (chromatin interaction analysis using paired end tag sequencing). In addition, global mRNA expression is also profiled with deep sequencing approach (RNA-seq)(84) with many cell types including T cells (85, 86).
Table 2.
Genome-wide approaches to analyze global epigenetic status
| Biological information |
Sequencing format |
Pre-sequencing sample preparation |
Description of genome-wide data | T cell data |
|---|---|---|---|---|
| Histone tail modification |
ChIP-seq | Chromatin immunoprecipitation |
Histone epigenome | Barski (Cell, 2007), Wei (Immunity, 2009, 2010), |
| Transcription factor binding |
ChIP-seq | Chromatin immunoprecipitation |
Transcription factor binding map |
Wei (Immunity, 2010)
Durant (Immunity 2010) |
| Nucleosome positioning |
MNase-seq | Micrococcal nuclease digestion |
Nucleosome landscape |
Valouev (Nature) 2011, Schones (Cell, 2008) |
| DNA accessibility | DNase-seq | DNase digestion | Chromatin accessibility map | Boyle (Cell, 2008) |
| DNA methylation | BS-seq MeDIP-seq, MBD-seq, MetylCap- seq |
Bisulfite conversion of cytosine Immunopresipitation Methy-CpG-binding domain |
DNA methylome | Deaton (Genome Research, 2011) |
| RNA expression | RNA-seq | mRNA | transcriptome |
Barski (Nature Struct. Bio., 2010), Hebenstreit (Mol. Sys. Bio. 2011) |
| miRNA expression |
miRNA-seq | miRNA | miRNAome | Kuchen (Immunity, 2010), Rossi (Nature Immunoloty, 2011) |
| Chromosome conformation |
4C-, 5C-, Hi-C-seq |
Chromatin proximity ligation |
Chromatin looping, globule, compartment | NA |
| Chromatin interaction through DNA binding protein |
ChIA-PET- seq |
ChIP + chromatin proximity ligation + paired end tags |
interactome | NA |
ChIP-seq for histone modification and transcription factor binding
This method allows for detection of protein-DNA interaction and therefore widely applied for profiling histone tail modifications, distribution of chromatin associated protein and binding of transcription factors (87). The technique involves crosslinking of protein to the contacting DNA and subsequent immunoprecipitation of DNA-protein complex by antibodies directed at a target protein. The pool of fragmented DNA enriched through immunoprecipitation is massively sequenced to produce a genome-wide digital map of local read frequencies that represent both location and strength of DNA-protein interaction.
MNase-seq for nucleosome positioning
This method employs micrococcal nuclease that preferentially cleaves DNA between nucleosomes (linker region DNA bridging juxtaposed nucleosomes or nucleosome free regions), thereby generates mononucleosomes preparation of approximately 200 bp DNA with associated core histones. By reading DNA sequences from the free end, the positions of individual nucleosomes can be mapped on a reference genome (88). Nucleosome-phasing surrounding the transcription start site of genes are reproducibly identified with notable differences between active and silent genes. Signal induced global remodeling of nucleosome organization was also captured by this analysis. In contrast, there are other genomic regions where the positioning of nucleosomes is substantially flexible and reproducibly positioned nucleosomes are seen for a small fraction. Therefore both DNA sequence-based nucleosome preference and non-nucleosomal factors are in action to determine nucleosome organization in mammalian cells (42).
DNase-seq for DNA accessibility
This method uses DNAse I to cut DNA within chromatin that is open and accessible for the enzyme, either away from nucleosome compacted area or not tightly covered by DNA associated proteins. Conventionally the positions of frequent cutting are referred to as DNAse hypersensitivity sites, and usually correspond to gene promoters or intronic as well as intergenic regulatory regions. Genomic sequencing of free ends generated by DNAse-mediated cutting identified hypersensitivity sites across the genome, in particular for distal intergenic regulatory elements with open chromosome configuration (89, 90).
Bisulfite-seq(MethylC-seq) to evaluate DNA methylation
This method relies on the nature of bisulfite treatment to selectively convert unmethylated cytosine to uracil so that subsequent deep sequencing would distinguish differential methylation status on a given cytosine residue at a single nucleotide resolution (34, 91). Because of high-resolution as well as lack of procedure-related bias, bisulfite-seq is the gold standard to evaluate global methylomes, despite its significantly high costs. Several alternative methods to profile DNA methylation such as ChIP based MeDIP-seq or methylated DNA binding domain sequencing are available with less cost but also with some compromises in resolution, sensitivity or coverage (92). Nevertheless they are useful applications when the investigation is focused on a selected region of the genome and genes and/or comparative analysis of multiple samples such as clinical samples (93) is required.
4C-seq and Hi-C-seq to evaluate chromosome conformation
The original technique to evaluate three dimensional chromosome architecture was named Chromosome Conformation Capture (3C) that generates proximity-based DNA ligation between 2 contacting chromosomal regions and subsequent quantitative detection of the ligation product by PCR (94). Original procedure was designed to detect interaction between a priori selected bait region and target region, but soon the technique has evolved to include many adaptations such as 4C (Circular Chromosome Conformation Capture)(95)(requiring a bait, but no prior knowledge on interacting regions), 5C (3C-Carbon Copy)(96)(high throughput 3C with multiplexed primers to cover ~400 kb wide region of choice), 3C-DSL(3C with DNA selection and ligation)(97)(5C coupled with IP for increased sensitivity and specificity) and Hi-C (98)(free of bias without prior selection of bait or target loci). Among those 3D assays, 4C and Hi-C are capable of providing genome-scale information when coupled with next generation sequencing. With Hi-C analysis that gives a resolution of 1 megabase, spatial segregation of open versus closed chromatin was observed as separate compartments, confirming the presence of chromosome territories.
ChIA-PET-seq for chromatin interaction through DNA binding protein
Ruan and colleagues has recently introduced a new strategy for whole genome chromatin interaction analysis by combining ChIP, chromatin proximity-detection by ligation, paired-end tag and high throughput sequencing (ChIA-PET; Chromatin interaction analysis using paired end tag sequencing)(99).
The basis of ChIA-PET is to introduce a linker sequence in the junction of two DNA fragments during nuclear proximity ligation to build connectivity of DNA fragments that are tethered together by protein factors (100). Subsequently all linker-connected ligation products are extracted as tag-linker-tag constructs that can be analyzed by high-throughput PET sequencing. Since chromatin interactions captured by chromatin proximity ligation are screened with chromatin IP, and not by any specific bait, ChIA-PET is an unbiased genome-wide approach for de novo detection of chromatin interactions mediated by a specific protein, collectively named as interactosome (100).
Using the ChIA-PET technique, the first chromatin interaction network was generated for the oestrogen receptor α (ER-α) in human genome. It was found that most high-confidence remote ER-α-binding sites are anchored at gene promoters through long-range chromatin interactions, suggesting that ER-α functions by extensive chromatin looping to bring genes together for coordinated transcriptional regulation (99).
Recently, the global CTCF-associated chromatin interactome map with high resolution has been generated for mouse embryonic stem (ES) cells (101). These results suggest that CTCF configures the genome into distinct chromatin domains and subnuclear compartments that exhibit unique epigenetic states and diverse transcriptional activities. Contrary to the enhancer-blocking model, CTCF-associated interactions potentially promote communications between functional regulatory elements to regulate gene expression. It was suggested that individual CTCF loops can function as domain barriers by demarcating nuclear lamin–chromatin interactions and delineating the chromosomes’ subnuclear localizations (101). Chromatin interactome map for T cells and its subsets are not reported yet.
Impact of epigenomic approaches to the understanding of Th differentiation
Comparative histone epigenomic profiling of Th subsets
Genome-wide H3K4 (active) and H3K27 (silent) trimethylation maps in naïve CD4+ T cells and fully polarized Th1, Th2, Th17, induced Treg and natural Treg cells have now been reported (102). These data allowed mapping of active, silent and poised chromatin states throughout the genome, and an interesting picture emerged. For genes encoding Th cell subset signature cytokines, the pattern is what would be expected - active marks are selectively present in the relevant lineage while silent marks are present in lineages that do not express the cytokine. In contrast, genes encoding “master regulator” transcription factors, such as Tbx21 in Th1 cells and Gata3 in Th2 cells, a more complex picture is evident. They do not exhibit marks associated with complete repression in the lineages in which they are not expressed but rather have “bivalent poised domains”, meaning both accessible and repressive marks are present (102). Bivalent domains were also present on genes encoding other key transcription factors including Runx3, Bcl6 and Blimp1 and provide a potential mechanism for flexibility in expression (102). Thus, histone epigenetic analysis provided evidences for both terminal commitment (e.g. cytokine genes) and plasticity (e.g. master regulators genes) of T helper cells, which coexist in the same lineage.
Effect of transcription factors on epigenetics and Th1/Th2 helper fate decision
Gene targeting of transcription factors that control lineage commitment provides the opportunity to determine how such factors regulate T helper fate, but importantly to also assess how they impact epigenetic processes (103, 104). While previous approaches have interrogated the effect of deletion of these transcription factors on specific genomic segments, global view of their actions can now be obtained.
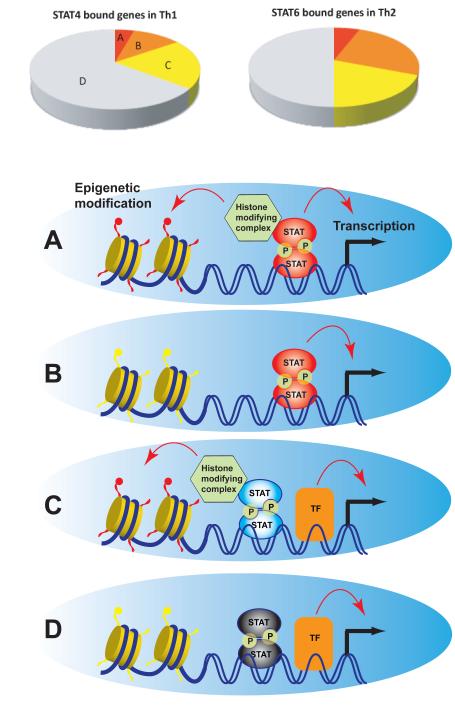
In the case of STAT4, 4 different categories of STAT4 action can be recognized (Fig.2) through comparative analysis of wild type- versus STAT4 knock out cells (104). Among the ~4000 genes directly bound by STAT4 (Fig.2 upper left pie chart), a minority (4%) are dependent on STAT4 for transcription competence and local histone epigenetic marks (H3K4me3, H3K27me3 or H3K36me3)(Fig.2-A). A subset (11%) is dependent on STAT4 for transcription but not for epigenetic modification (Fig. 2-B), with a larger proportion (20%) being dependent upon STAT4 for epigenetic modificaiton but not for transcription (Fig. 2-C). The majority (64%) are neither dependent on STAT4 for transcription nor epigenetic changes despite that STAT4 binds to the genes (Fig. 2-D). Similar analysis for STAT6 revealed even more profound global effect of STAT6 on epigenetic marks in Th2 conditions (Fig. 2 upper right pie chart). These observations indicate that there are multiple different ways a transcription factor can deliver its effect, and that effects on epigenetic processes represent a significant aspect of its action on a genome-wide scale.
Fig.2.
- Both transcription and local epigenetic status are dependent on STAT
- Transcription is STAT-dependent but local epigenetic status is not
- Epigenetic status is STAT-dependent but gene transcription is not
- Neither transcription nor epigenetic status are STAT-dependent
Dual function of transcription factor: activator and repressor
In general, transcription factors drive lineage commitment by both positively and negatively through promoting the expression of phenotype-defining genes on one hand and repressing the expression of genes associated with alternative fates on the other hand. Although STATs were originally discovered as activators of gene transcription, a point that has been well-confirmed by genome-wide analysis, there have been few indications that STATs can also function as direct repressors (105, 106). In T cells, STAT4-dependent repressive histone marks were identified on several Th2 cell-expressed genes, including STAT6 targets, which are actively repressed by STAT4 in Th1 cells (104). In total, around 40 genes are identified as having STAT4-dependent repressive histone marks, and suggesting a role for STAT4 as a transcriptional repressor as well as its more widely recognized role as a transcriptional activator. The successful identification of opposing chromatin modifications associated with STAT binding illustrates the power of genomic approach to evaluate differential mode of transcription factor, both positive and negative, on the genome-wide scale.
Epigenetic instability of Th17 cells
Analogous to Th1 or Th2 cells, the defining phenotype of Th17 cells relates to the expression of IL-17 and master regulator Rorc. However, unlike Th1 and Th2 cells, there is considerable intrinsic instability of Th17. Extended culture of Th17 cells in vitro or upon adoptive transfer can result in loss of IL-17 production and acquisition of IFN-γ production (107-109). Upon exposure to IL-12, IFN-γ production can be rapidly induced in Th17 producing cells with down-regulation of Rorc and extinction of IL-17 production (107). It was noted that significant remodeling of distal enhancers in the Ifng locus was already present in Th17 cells, even in the absence of Th1-specifying cytokines, a factor thought to be important in allowing rapid induction of IFN-γ (107). In contrast, the Il17a/f locus in Th1 cells is devoid of signs of remodeling in Th1 cells, suggesting asymmetry in the relationship between Th1 and Th17 cells. Maintenance of IL-17 production requires continued expression of Rorc provided through STAT3 activating signals such as IL-6, IL-23 and IL-21 and inhibition of expression of T-bet and GATA3 (1, 107). In Th17 cells, STAT3 binds not only Rorc but other transcription factors and cytokine receptors associated with the Th17 phenotype, many of which also carry active epigenetic marks on their promoters (103).
While STAT3 has a direct positive role in promoting Th17 differentiation, it appears that STAT5 has a direct negative role. It is well established that IL-2 inhibits Th17 differentiation and by acting as IL-2 “sinks” Treg cells can promote Th17 differentiation (110-112). Using genome-wide ChIP-seq analysis, it has noted that STAT3 and STAT5 bound the same binding sites in the Il-17a-IL17f locus in CD4+ T cells (113). It was found that IL-2-induced STAT5 binding was associated with a reduction of STAT3 binding and also a reduction of positive epigenetic marks at multiple sites throughout the Il-17a-Il17f locus in Th17 cells. This is yet another example in which STAT proteins can repress gene expression, in this case not just by promoting the accumulation of repressive marks, but also by directly competing with a positive regulator of gene transcription.
TGF-β is another factor that promotes Th17 differentiation. One way it seems to work is by dampening expression of T-bet, GATA3, IFN-γ and other factors that allow Th17 cells to drift to become Th1 cells (109). It should be noted that Th17 cells can be generated in the absence of TGF-β and cells generated in this manner express Rorc and T-bet (114, 115).
Plasticity of Tfh cells and insights from epigenetic analysis
Tfh cells have recently been proposed as another lineage with specialized function in promoting B cell mediated antibody responses. Tfh cells are defined by expression of master regulator Bcl-6 and effector cytokine IL-21, in addition to their localization in germinal center of lymphoid tissues. A pattern of gene expression unique to Tfh is also reported which include expression of the following surface receptors: CXCR5, PD-1, ICOS, BTLA, and CD84. However substantial plasticity between Tfh and other effector cell populations is also recognized, supporting the concept of reprogramming plasticity of Tfh. Tfh cells may arise from Treg and Th2 cells and conversely, Tfh may differentiate to become Th1 and other effector cells (9).
Global epigenetic profiling of in vitro generated- and in vivo-derived Tfh has provided additional evidences to support plasticity of Tfh cells and involvement of epigenetic regulation of Bcl6. Of note was that the Bcl6 locus maintains accessible epigenetic marks, even in polarized effector subsets to a degree that is comparable to Tfh (Lu et al Immunity in press). This argues that this key transcription factor may be readily induced – even in cells that appear to be firmly committed to being Th1, Th2, and Th17 cells.
Recent understanding on genomic structure outside of annotated genes
Enhancers
As illustrated by cytokines, tight regulation of gene expression is controlled by enhancer elements, which can reside many kilobases distal from the genes they regulate. Until recently, evidence of enhancer function relied primarily on conservation (and therefore denoted conserved noncoding sequences or CNS) and DNase hypersensitivity sites (116, 117).
However, sequence conservation does not provide any information on the tissue(s) where that enhancer is actually active and also conservation of regulatory function is not always faithfully associated with the conservation of DNA sequence. The advent of next generation sequencing has provided an “enhancer signaure” to look for, namely monomethylation of histone H3 lysine 4 (H3K4me1) in the absence of trimetylation (H3K4me3) (118-120) and binding of p300 histone acetyltransferase (116, 121-123). The veracity of enhancers identified in this manner has been established through the construction of transgenic reporter mice, which successfully reproduced tissue-specific and developmentally timed gene expression (123). In addition, combinatorial patterns of histone epigenetic marks on enhancer elements can distinguish the state of enhancer activity as being active, poised or repressed. That is, acetylation of histone H3K27 in combination with H3K4me1 mark active enhancers, whereas H3K4me1, in the absence of H3K27 acetylation, mark inactive or “poised” enhancers (124, 125). Many poised enhancers in ES cells were associated with repressive H3K27me3, and some were replaced by active H3K27ac upon differentiation to a neuronal pathway (125). Collectively, H3K4me1 represents a generalized mark for distal enhancers and additional marks distinguish the state of enhancer activity (116). Until recently, our understanding of the enhancer structure has been limited to very few genes; however, these new approaches provide the opportunity to vastly expand our understanding on the structure and function of regulatory elements. Considering how much of the genome corresponds to non-coding regions or “gene desert”, it is intriguing that disease-associated polymporphisms are now being linked to such regions (97, 126). For instance, by connecting enhancer chromatin signatures and transcription factor binding sites, one noncoding region (9p21) associated with coronary artery disease was recognized to be functionally related to impaired IFN-γ signaling. This represents just one example of how new strategies can begin to decipher biological functions of loci in “gene deserts”.
Insulators
Classically, insulators are defined by the ability to block communication between adjacent regulatory elements in a position-dependent manner and the capacity to buffer transgenes by the spread of repressive heterochromatin from adjacent sequences. The binding of CTCF (CCCTC-binding factor) and its partners is typically associated with the presence of insulator elements, and the boundaries of the Ifng and IFNG loci are associated CTCF binding (54, 55). Based on the recruitment of CTCF/Rad21, the murine Ifng locus extends from −70 to +66 kb relative to the transcription start site (55, 57). Recent work points to more complex functions of CTCF and its partners (e.g. cohesin). These factors are recognized as having a role in three-dimensional chromatin conformation and a genome-wide role in the organization of developmentally regulated intra- and interchromosomal contacts (127, 128). In this light, the traditional regulatory functions of CTCF such as transcriptional activation, repression, insulation, and imprinting, may all be secondary effects of its primary role as a genome-wide organizer of chromatin architecture (101, 128).
Roles of long and short non-coding RNA
Transcriptional and epigenetic regulation is generally described as dynamic processes mediated by a protein network. Recently, however, it has become increasingly apparent that there is an additional layer of regulation mediated not by protein, but by non-protein-coding RNA (ncRNA). The mammalian genome is pervasively transcribed over a variety of cell types with the majority of the transcription composed of ncRNA (118, 129) which include microRNA, enhancer RNA (130) and long non-coding RNA.
miRNA
MiRNAs are small, ~22 nucleotide single-stranded ncRNAs that mediate post-transcriptional regulation of transcriptional networks. Many reviews have covered this exciting area (131, 132) and therefore this current review will focus discussion on the function of miRNA in T cells. Drosha and Dicer are two key components of the machinery responsible for miRNA generation (133) and with respect to T cells, loss of these factors is associated with instability of helper T cells and consequently autoimmunity (134-136).
In order to identify critical miRNAs that drive T helper cell differentiation and lineage commitment, comprehensive atlases of the murine and human lymphocyte ‘microRNome’ were created (137, 138). Using RNA-seq and RT-qPCR to analyze miRNA expression in over 50 cell types between the two studies, it was found that 49 (of 600 screened; ~8%) murine and 29 (of 664 screened; ~4%) human miRNAs were preferentially and differentially upregulated in lymphocytes. Identification of miRNAs whose expression was confined to specific lineage might imply their unique roles in regulating lineage specificity (139, 140). Several miRNAs that influence T helper cell function have been reported. miR-125b contributes to enforcing naïve state of helper T cells (138). miR-182 promotes clonal expansion (141). miR-326 was reported to enhance TH17 differentiation (142). miR-146a is important for suppressive function of Treg (143).
With miRNAs as key modulators of RNA stability and lncRNAs emerging as potent transcriptional regulators, ncRNAs have demonstrated that they are significant regulatory players within the transcriptional network. This paradigm will likely be underscored even more in the near future with the recent introduction of enhancer RNAs (eRNAs)(130) and promoter-associated RNAs (144, 145) to ncRNA repertoire.
Long non-coding RNA (lncRNA)
LncRNAs are a remarkably diverse class of transcripts that together are defined as being larger than 200 nucleotides and lacking a functional open reading frame. LncRNAs may be 5′ capped, polyadenylated, and originate from intron/exon architecture that may be alternatively spliced. LncRNAs are found throughout the genome, having been identified within intergenic regions and intragenically, where they overlap with known protein-coding genes or other ncRNAs in an anti-sense or sense orientation (146, 147). A large number of lncRNAs have been identified in a wide variety of cell types using a combination of tiling arrays, RNA-seq, large-scale cDNA sequencing, and the presence of H3K4me3 and H3K36me3 histone modifications (148).
While relatively few lncRNAs have been characterized for their function, lncRNAs appear to play a prominent role forming ribonucleoprotein complexes that mediate epigenetic regulation and transcriptional expression of highly specified target genes (149). For instance, the lncRNA HOTAIR acts as a scaffold for the coordinated recruitment of both PRC2 and the CoREST/REST repressor complex for coupled induction of H3K27 methylation and H3K4 demethylation on the HOXD locus (150). Similarly, HOTTIP mediates the recruitment of WDR5 to target genes driving H3K4 trimethylation and transcriptional activation (151).
Not only RNA is an integral component of chromatin but also many histone methyltransferases lack DNA binding properties and instead possessing RNA binding motifs, indicating that lncRNAs may play a much more pervasive role than currently recognized (152). In T cells, a few lncRNAs that have been functionally characterized include: NRON (ncRNA repressor of the nuclear factor of activated T cells)(153, 154), GAS5 (growth arrest specific transcript 5)(155) and lncRNA from the T early alpha promoter (TEA)(156, 157). Clearly though, this is an area that will explode over the next few years.
Immunological disease and epigenetics
The pathological significance of epigenetic gene regulation has been very well appreciated in oncology, but increasingly such mechanisms are being recognized as contributors to various immunological disorders. This makes sense insofar as environmental triggers play an important role in such diseases. Among them, we will highlight emerging data on the contribution of epigenetic mechanisms to two diseases, systemic lupus erythematosus (SLE) and asthma.
SLE is an autoimmune disease of unknown etiology with genetic, hormonal and environmental influences. The idea that epigenetic mechanism plays a role in lupus stemmed from initial observations that medications that inhibited DNA methylation could trigger the disease (158). In addition, while studies on twins pointed to genetic factors to this disease, incomplete concordance pointed to other factors as well. In this regard, it has been recognized for more than two decades that CD4+ T cells from patients with SLE exhibited hypomethylated DNA. Importantly, this difference has been found in twins discordant for lupus. In addition, T cells and monocytes from lupus patients have also been noted to have alterations in histone modifications (159).
Like SLE, asthma clearly has a genetic component; however, the recent rise in the incidence of this disease cannot be explained only by genetics. As discussed in detail throughout this review and others, the expression of cytokine genes and other loci, which are relevant to the pathogenesis of asthma, are highly influenced by epigenetic mechanisms. Environmental factors such as tobacco smoke and pollutants also influence DNA methylation and histone modifications and other chromatin modifications (160).
Conclusion/future perspectives
It should be quite evident that new technologic approaches have vastly expanded our view on cell phenotype and we now think at the level of “-ome”s - epigenome, transcriptome, miRNAome, methylome, interactome and proteosome to define cellular phenotype. Integrating volumes of “omic”-information and making sense of these data will be a real challenge. Linking such information to GWAS studies and the predisposition to human diseases has already yielded promising mechanistic clues to better understand their pathophysiology. Future studies will certainly continue to extract more information hidden in “gene desert” regions of the genome. Comprehensively understanding the dynamic interactions between the genome and epigenome will revolutionize our view on how T cell function in health and disease.
Equally, the epigenome is recognized as being dynamically shaped by signals in the microenvironment. In this respect, factors that regulate epigenetic changes are increasingly recognized as an extension of signal transduction mechanisms. The mechanisms through which transcription factors recruit chromatin modifying enzymes to organize epigenome remains poorly understood; however, there is no doubt that we will learn much in the next few years about how nucleosome biology is linked biochemically with signaling transduction. This will provide us with a much more sophisticated view of lymphocyte cell biology, activation and the control of gene expression.
References
- 1.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nature Reviews Immunology. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 2.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations. Annu Rev Immunol. 2010;28:445–89. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ansel KM, Greenwald RJ, Agarwal S, Bassing CH, Monticelli S, Interlandi J, Djuretic IM, Lee DU, Sharpe AH, Alt FW, Rao A. Deletion of a conserved Il4 silencer impairs T helper type 1-mediated immunity. Nat Immunol. 2004;5:1251–9. doi: 10.1038/ni1135. [DOI] [PubMed] [Google Scholar]
- 4.Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annual Review of Immunology. 2006;24:607–56. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 5.Crotty S. Annual Review of Immunology. 2011. Follicular Helper CD4 T cells (TFH) pp. 621–63. [DOI] [PubMed] [Google Scholar]
- 6.Eyerich S, Eyerich K, Pennino D, Carbone T, Nasorri F, Pallotta S, Cianfarani F, Odorisio T, Traidl-Hoffmann C, Behrendt H, Durham SR, Schmidt-Weber CB, Cavani A. Th22 cells represent a distinct human T cell subset involved in epidermal immunity and remodeling. J Clin Invest. 2009;119:3573–85. doi: 10.1172/JCI40202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veldhoen M, Uyttenhove C, van Snick J, Helmby H, Westendorf A, Buer J, Martin B, Wilhelm C, Stockinger B. Transforming growth factor-beta ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat Immunol. 2008;9:1341–6. doi: 10.1038/ni.1659. [DOI] [PubMed] [Google Scholar]
- 8.Chang HC, Sehra S, Goswami R, Yao W, Yu Q, Stritesky GL, Jabeen R, McKinley C, Ahyi AN, Han L, Nguyen ET, Robertson MJ, Perumal NB, Tepper RS, Nutt SL, Kaplan MH. The transcription factor PU.1 is required for the development of IL-9-producing T cells and allergic inflammation. Nat Immunol. 2010;11:527–34. doi: 10.1038/ni.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Shea J, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4 + T cells. Science. 2010;327:1098–102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hegazy AN, Peine M, Helmstetter C, Panse I, Fr√∂hlich A, Bergthaler A, Flatz L, Pinschewer DD, Radbruch A, Lohning M. Interferons Direct Th2 Cell Reprogramming to Generate a Stable GATA-3+T-bet+ Cell Subset with Combined Th2 and Th1 Cell Functions. Immunity. 2010;32:116–28. doi: 10.1016/j.immuni.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 11.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–94. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- 12.Hwang ES, Szabo SJ, Schwartzberg PL, Glimcher LH. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science. 2005;307:430–3. doi: 10.1126/science.1103336. [DOI] [PubMed] [Google Scholar]
- 13.Amsen D, Blander JM, Lee GR, Tanigaki K, Honjo T, Flavell RA. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell. 2004;117:515–26. doi: 10.1016/s0092-8674(04)00451-9. [DOI] [PubMed] [Google Scholar]
- 14.Kim JI, Ho IC, Grusby MJ, Glimcher LH. The transcription factor c-Maf controls the production of interleukin-4 but not other Th2 cytokines. Immunity. 1999;10:745–51. doi: 10.1016/s1074-7613(00)80073-4. [DOI] [PubMed] [Google Scholar]
- 15.Stritesky GL, Muthukrishnan R, Sehra S, Goswami R, Pham D, Travers J, Nguyen ET, Levy DE, Kaplan MH. The Transcription Factor STAT3 Is Required for T Helper 2 Cell Development. Immunity. 2011;34:39–49. doi: 10.1016/j.immuni.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu Q, Sharma A, Ghosh A, Sen JM. T cell factor-1 negatively regulates expression of IL-17 family of cytokines and protects mice from experimental autoimmune encephalomyelitis. J Immunol. 2011;186:3946–52. doi: 10.4049/jimmunol.1003497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou L, Lopes JE, Chong MM, Ivanov II, Min R, Victora GD, Shen Y, Du J, Rubtsov YP, Rudensky AY, Ziegler SF, Littman DR. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–40. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang F, Meng G, Strober W. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat Immunol. 2008;9:1297–306. doi: 10.1038/ni.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rudra D, Egawa T, Chong MM, Treuting P, Littman DR, Rudensky AY. Runx-CBFbeta complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat Immunol. 2009;10:1170–7. doi: 10.1038/ni.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sekiya T, Kashiwagi I, Inoue N, Morita R, Hori S, Waldmann H, Rudensky AY, Ichinose H, Metzger D, Chambon P, Yoshimura A. The nuclear orphan receptor Nr4a2 induces Foxp3 and regulates differentiation of CD4+ T cells. Nat Commun. 2011;2:269. doi: 10.1038/ncomms1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerdiles YM, Stone EL, Beisner DR, McGargill MA, Ch’en IL, Stockmann C, Katayama CD, Hedrick SM. Foxo transcription factors control regulatory T cell development and function. Immunity. 2010;33:890–904. doi: 10.1016/j.immuni.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng Y, Chaudhry A, Kas A, deRoos P, Kim JM, Chu TT, Corcoran L, Treuting P, Klein U, Rudensky AY. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–6. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, Rudensky AY. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–91. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beyer M, Thabet Y, Muller RU, Sadlon T, Classen S, Lahl K, Basu S, Zhou X, Bailey-Bucktrout SL, Krebs W, Schonfeld EA, Bottcher J, Golovina T, Mayer CT, Hofmann A, Sommer D, Debey-Pascher S, Endl E, Limmer A, Hippen KL, Blazar BR, Balderas R, Quast T, Waha A, Mayer G, Famulok M, Knolle PA, Wickenhauser C, Kolanus W, Schermer B, Bluestone JA, Barry SC, Sparwasser T, Riley JL, Schultze JL. Repression of the genome organizer SATB1 in regulatory T cells is required for suppressive function and inhibition of effector differentiation. Nat Immunol. 2011;12:898–907. doi: 10.1038/ni.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waddington CH. An introduction to modern genetics. The macmillan company. 1939 [Google Scholar]
- 27.Smith E, Lin C, Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 2011;25:661–72. doi: 10.1101/gad.2015411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–9. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem. 2010;79:155–79. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- 30.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peserico A, Simone C. Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J Biomed Biotechnol. 2011;2011:371832. doi: 10.1155/2011/371832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–31. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Talbert PB, Henikoff S. Histone variants--ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11:264–75. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 34.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O’Malley R, Castanon R, Klugman S, Downes M, Yu R, Stewart R, Ren B, Thomson JA, Evans RM, Ecker JR. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. Chembiochem. 2011;12:206–22. doi: 10.1002/cbic.201000195. [DOI] [PubMed] [Google Scholar]
- 37.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–20. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pastor WA, Pape UJ, Huang Y, Henderson HR, Lister R, Ko M, McLoughlin EM, Brudno Y, Mahapatra S, Kapranov P, Tahiliani M, Daley GQ, Liu XS, Ecker JR, Milos PM, Agarwal S, Rao A. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–7. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szulwach KE, Li X, Li Y, Song CX, Han JW, Kim S, Namburi S, Hermetz K, Kim JJ, Rudd MK, Yoon YS, Ren B, He C, Jin P. Integrating 5-hydroxymethylcytosine into the epigenomic landscape of human embryonic stem cells. PLoS Genet. 2011;7:e1002154. doi: 10.1371/journal.pgen.1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–3. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bai L, Morozov AV. Gene regulation by nucleosome positioning. Trends in Genetics. 2010;26:476–83. doi: 10.1016/j.tig.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 42.Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–22. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bell O, Tiwari VK, Thomä NH, Schábeler D. Determinants and dynamics of genome accessibility. Nature Reviews Genetics. 2011 doi: 10.1038/nrg3017. [DOI] [PubMed] [Google Scholar]
- 44.Chi T. A BAF-centred view of the immune system. Nat Rev Immunol. 2004;4:965–77. doi: 10.1038/nri1501. [DOI] [PubMed] [Google Scholar]
- 45.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–84. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cairns BR. Chromatin remodeling complexes: Strength in diversity, precision through specialization. Current Opinion in Genetics and Development. 2005;15:185–90. doi: 10.1016/j.gde.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 47.Chi TH, Wan M, Zhao K, Taniuchi I, Chen L, Uttman DR, Crabtree GR. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature. 2002;418:195–9. doi: 10.1038/nature00876. [DOI] [PubMed] [Google Scholar]
- 48.Deng W, Blobel GA. Do chromatin loops provide epigenetic gene expression states? Curr Opin Genet Dev. 2010;20:548–54. doi: 10.1016/j.gde.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nature Immunology. 2004;5:1017–27. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- 50.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–45. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 51.Sutherland H, Bickmore WA. Transcription factories: gene expression in unions? Nat Rev Genet. 2009;10:457–66. doi: 10.1038/nrg2592. [DOI] [PubMed] [Google Scholar]
- 52.Aune TM, Collins PL, Chang S. Epigenetics and T helper 1 differentiation. Immunology. 2009;126:299–305. doi: 10.1111/j.1365-2567.2008.03026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu H, Yang J, Murphy TL, Ouyang W, Wagner F, Saparov A, Weaver CT, Murphy KM. Unexpected characteristics of the IFN-gamma reporters in nontransformed T cells. Journal of Immunology. 2001;167:855–65. doi: 10.4049/jimmunol.167.2.855. [DOI] [PubMed] [Google Scholar]
- 54.Hadjur S, Williams LM, Ryan NK, Cobb BS, Sexton T, Fraser P, Fisher AG, Merkenschlager M. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460:410–U130. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sekimata M, Perez-Melgosa M, Miller SA, Weinmann AS, Sabo PJ, Sandstrom R, Dorschner MO, Stamatoyannopoulos JA, Wilson CB. CCCTC-Binding Factor and the Transcription Factor T-bet Orchestrate T Helper 1 Cell-Specific Structure and Function at the Interferon-gamma Locus. Immunity. 2009;31:551–64. doi: 10.1016/j.immuni.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Balasubramani A, Shibata Y, Crawford GE, Baldwin AS, Hatton RD, Weaver CT. Modular Utilization of Distal cis-Regulatory Elements Controls Ifng Gene Expression in T Cells Activated by Distinct Stimuli. Immunity. 2010;33:35–47. doi: 10.1016/j.immuni.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Balasubramani A, Mukasa R, Hatton RD, Weaver CT. Regulation of the Ifng locus in the context of T-lineage specification and plasticity. Immunological Reviews. 2010;238:216–32. doi: 10.1111/j.1600-065X.2010.00961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee GR, Fields PE, Griffin Iv TJ, Flavell RA. Regulation of the Th2 cytokine locus by a locus control region. Immunity. 2003;19:145–53. doi: 10.1016/s1074-7613(03)00179-1. [DOI] [PubMed] [Google Scholar]
- 59.Kim ST, Fields PE, Flavell RA. Demethylation of a specific hypersensitive site in the Th2 locus control region. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:17052–7. doi: 10.1073/pnas.0708293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Djuretic IM, Levanon D, Negreanu V, Groner Y, Rao A, Ansel KM. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat Immunol. 2007;8:145–53. doi: 10.1038/ni1424. [DOI] [PubMed] [Google Scholar]
- 61.Naoe Y, Setoguchi R, Akiyama K, Muroi S, Kuroda M, Hatam F, Littman DR, Taniuchi I. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf beta binding to the Il4 silencer. J Exp Med. 2007;204:1749–55. doi: 10.1084/jem.20062456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, Schlawe K, Chang HD, Bopp T, Schmitt E, Klein-Hessling S, Serfling E, Hamann A, Huehn J. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biology. 2007;5:0169–78. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim HP, Leonard WJ. CREB/ATF-dependent T cell receptor-induced FoxP3 gene expression: a role for DNA methylation. J Exp Med. 2007;204:1543–51. doi: 10.1084/jem.20070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huehn J, Polansky JK, Hamann A. Epigenetic control of FOXP3 expression: the key to a stable regulatory T-cell lineage? Nat Rev Immunol. 2009;9:83–9. doi: 10.1038/nri2474. [DOI] [PubMed] [Google Scholar]
- 65.Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–12. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ballas ZK. The use of 5-azacytidine to establish constitutive interleukin 2-producing clones of the EL4 thymoma. J Immunol. 1984;133:7–9. [PubMed] [Google Scholar]
- 67.Young HA, Ghosh P, Ye J, Lederer J, Lichtman A, Gerard JR, Penix L, Wilson CB, Melvin AJ, McGurn ME, Lewis DB, Taub DD. Differentiation of the T helper phenotypes by analysis of the methylation state of the IFN-Œ≥ gene. Journal of Immunology. 1994;153:3603–10. [PubMed] [Google Scholar]
- 68.Bird JJ, Brown DR, Mullen AC, Moskowitz NH, Mahowald MA, Sider JR, Gajewski TF, Wang CR, Reiner SL. Helper T cell differentiation is controlled by the cell cycle. Immunity. 1998;9:229–37. doi: 10.1016/s1074-7613(00)80605-6. [DOI] [PubMed] [Google Scholar]
- 69.Valapour M, Guo J, Schroeder JT, Keen J, Cianferoni A, Casolaro V, Georas SN. Histone deacetylation inhibits IL4 gene expression in T cells. Journal of Allergy and Clinical Immunology. 2002;109:238–45. doi: 10.1067/mai.2002.121145. [DOI] [PubMed] [Google Scholar]
- 70.Morinobu A, Kanno Y, O’Shea JJ. Discrete roles for histone acetylation in human T helper 1 cell-specific gene expression. Journal of Biological Chemistry. 2004;279:40640–6. doi: 10.1074/jbc.M407576200. [DOI] [PubMed] [Google Scholar]
- 71.Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, P√©rez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, Cherry SR, Tsai JH, Tucker SM, Weaver WM, Kelso A, Jaenisch R, Wilson CB. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15:763–74. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 72.Makar KW, P√©rez-Melgosa M, Shnyreva M, Weaver WM, Fitzpatrick DR, Wilson CB. Active recruitment of DNA methyltransferases regulates interleukin 4 in thymocytes and T cells. Nature Immunology. 2003;4:1183–90. doi: 10.1038/ni1004. [DOI] [PubMed] [Google Scholar]
- 73.Hutchins AS, Mullen AC, Lee HW, Sykes KJ, High FA, Hendrich BD, Bird AP, Reiner SL. Gene silencing quantitatively controls the function of a developmental trans-activator. Molecular Cell. 2002;10:81–91. doi: 10.1016/s1097-2765(02)00564-6. [DOI] [PubMed] [Google Scholar]
- 74.Yamashita M, Hirahara K, Shinnakasu R, Hosokawa H, Norikane S, Kimura MY, Hasegawa A, Nakayama T. Crucial role of MLL for the maintenance of memory T helper type 2 cell responses. Immunity. 2006;24:611–22. doi: 10.1016/j.immuni.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 75.Onodera A, Yamashita M, Endo Y, Kuwahara M, Tofukuji S, Hosokawa H, Kanai A, Suzuki Y, Nakayama T. STAT6-mediated displacement of polycomb by trithorax complex establishes long-term maintenance of GATA3 expression in T helper type 2 cells. J Exp Med. 2010;207:2493–506. doi: 10.1084/jem.20100760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kimura M, Koseki Y, Yamashita M, Watanabe N, Shimizu C, Katsumoto T, Kitamura T, Taniguchi M, Koseki H, Nakayama T. Regulation of Th2 cell differentiation by mel-18, a mammalian Polycomb group gene. Immunity. 2001;15:275–87. doi: 10.1016/s1074-7613(01)00182-0. [DOI] [PubMed] [Google Scholar]
- 77.Yamashita M, Kuwahara M, Suzuki A, Hirahara K, Shinnaksu R, Hosokawa H, Hasegawa A, Motohashi S, Iwama A, Nakayama T. Bmi1 regulates memory CD4 T cell survival via repression of the Noxa gene. J Exp Med. 2008;205:1109–20. doi: 10.1084/jem.20072000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koyanagi M, Baguet A, Martens J, Margueron R, Jenuwein T, Bix M. EZH2 and histone 3 trimethyl lysine 27 associated with Il4 and Il13 gene silencing in TH1 cells. Journal of Biological Chemistry. 2005;280:31470–7. doi: 10.1074/jbc.M504766200. [DOI] [PubMed] [Google Scholar]
- 79.Su IH, Dobenecker MW, Dickinson E, Oser M, Basavaraj A, Marqueron R, Viale A, Reinberg D, Wulfing C, Tarakhovsky A. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell. 2005;121:425–36. doi: 10.1016/j.cell.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 80.Gebuhr TC, Kovalev GI, Bultman S, Godfrey V, Su L, Magnuson T. The Role of Brg1, a Catalytic Subunit of Mammalian Chromatin-remodeling Complexes, in T Cell Development. Journal of Experimental Medicine. 2003;198:1937–49. doi: 10.1084/jem.20030714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chi TH, Wan M, Lee PP, Akashi K, Metzger D, Chambon P, Wilson CB, Crabtree GR. Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity. 2003;19:169–82. doi: 10.1016/s1074-7613(03)00199-7. [DOI] [PubMed] [Google Scholar]
- 82.Zhang F, Boothby M. T helper type 1-specific Brg1 recruitment and remodeling of nucleosomes positioned at the IFN-Œ≥ promoter are Stat4 dependent. Journal of Experimental Medicine. 2006;203:1493–505. doi: 10.1084/jem.20060066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Landry JW, Banerjee S, Taylor B, Aplan PD, Singer A, Wu C. Chromatin remodeling complex NURF regulates thymocyte maturation. Genes and Development. 25:275–86. doi: 10.1101/gad.2007311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Barski A, Chepelev I, Liko D, Cuddapah S, Fleming AB, Birch J, Cui K, White RJ, Zhao K. Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. Nat Struct Mol Biol. 2010;17:629–34. doi: 10.1038/nsmb.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hebenstreit D, Fang M, Gu M, Charoensawan V, van Oudenaarden A, Teichmann SA. RNA sequencing reveals two major classes of gene expression levels in metazoan cells. Mol Syst Biol. 2011;7:497. doi: 10.1038/msb.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–80. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–98. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Crawford GE, Holt IE, Mullikin JC, Tai D, Blakesley R, Bouffard G, Young A, Masiello C, Green ED, Wolfsberg TG, Collins FS. Identifying gene regulatory elements by genome-wide recovery of DNase hypersensitive sites. Proc Natl Acad Sci U S A. 2004;101:992–7. doi: 10.1073/pnas.0307540100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sabo PJ, Humbert R, Hawrylycz M, Wallace JC, Dorschner MO, McArthur M, Stamatoyannopoulos JA. Genome-wide identification of DNaseI hypersensitive sites using active chromatin sequence libraries. Proc Natl Acad Sci U S A. 2004;101:4537–42. doi: 10.1073/pnas.0400678101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ji H, Ehrlich LI, Seita J, Murakami P, Doi A, Lindau P, Lee H, Aryee MJ, Irizarry RA, Kim K, Rossi DJ, Inlay MA, Serwold T, Karsunky H, Ho L, Daley GQ, Weissman IL, Feinberg AP. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467:338–42. doi: 10.1038/nature09367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Deaton AM, Webb S, Kerr ARW, Illingworth RS, Guy J, Andrews R, Bird A. Cell type-specific DNA methylation at intragenic CpG islands in the immune system. Genome Research. 21:1074–86. doi: 10.1101/gr.118703.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–9. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 94.Sanyal A, Ba√π D, Mart√≠-Renom MA, Dekker J. Chromatin globules: A common motif of higher order chromosome structure? Current Opinion in Cell Biology. 23:325–31. doi: 10.1016/j.ceb.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38:1341–7. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 96.Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, Green RD, Dekker J. Chromosome Conformation Capture Carbon Copy (5C): A massively parallel solution for mapping interactions between genomic elements. Genome Research. 2006;16:1299–309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Harismendy O, Notani D, Song X, Rahim NG, Tanasa B, Heintzman N, Ren B, Fu XD, Topol EJ, Rosenfeld MG, Frazer KA. 9p21 DNA variants associated with coronary artery disease impair interferon-gamma signalling response. Nature. 2011;470:264–8. doi: 10.1038/nature09753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lieberman-Aiden E, Van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, Chew EGY, Huang PYH, Welboren WJ, Han Y, Ooi HS, Ariyaratne PN, Vega VB, Luo Y, Tan PY, Choy PY, Wansa KDSA, Zhao B, Lim KS, Leow SC, Yow JS, Joseph R, Li H, Desai KV, Thomsen JS, Lee YK, Karuturi RKM, Herve T, Bourque G, Stunnenberg HG, Ruan X, Cacheux-Rataboul V, Sung WK, Liu ET, Wei CL, Cheung E, Ruan Y. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fullwood MJ, Ruan Y. ChIP-based methods for the identification of long-range chromatin interactions. Journal of Cellular Biochemistry. 2009;107:30–9. doi: 10.1002/jcb.22116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CWH, Ye C, Ping JLH, Mulawadi F, Wong E, Sheng J, Zhang Y, Poh T, Chan CS, Kunarso G, Shahab A, Bourque G, Cacheux-Rataboul V, Sung WK, Ruan Y, Wei CL. CTCF-mediated functional chromatin interactome in pluripotent cells. Nature Genetics. 43:630–8. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, Schones DE, Peng W, Sun Hw, Paul WE, O’Shea JJ, Zhao K. Global Mapping of H3K4me3 and H3K27me3 Reveals Specificity and Plasticity in Lineage Fate Determination of Differentiating CD4+ T Cells. Immunity. 2009;30:155–67. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Durant L, Watford WT, Ramos HL, Laurence A, Vahedi G, Wei L, Takahashi H, Sun HW, Kanno Y, Powrie F, O’Shea JJ. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 2010;32:605–15. doi: 10.1016/j.immuni.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wei L, Vahedi G, Sun HW, Watford WT, Takatori H, Ramos HL, Takahashi H, Liang J, Gutierrez-Cruz G, Zang CZ, Peng WQ, O’Shea JJ, Kanno Y. Discrete Roles of STAT4 and STAT6 Transcription Factors in Tuning Epigenetic Modifications and Transcription during T Helper Cell Differentiation. Immunity. 2010;32:840–51. doi: 10.1016/j.immuni.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Walker SR, Nelson EA, Frank DA. STAT5 represses BCL6 expression by binding to a regulatory region frequently mutated in lymphomas. Oncogene. 2007;26:224–33. doi: 10.1038/sj.onc.1209775. [DOI] [PubMed] [Google Scholar]
- 106.Tran TH, Utama FE, Lin J, Yang N, Sjolund AB, Ryder A, Johnson KJ, Neilson LM, Liu C, Brill KL, Rosenberg AL, Witkiewicz AK, Rui H. Prolactin inhibits BCL6 expression in breast cancer through a Stat5a-dependent mechanism. Cancer Res. 2010;70:1711–21. doi: 10.1158/0008-5472.CAN-09-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mukasa R, Balasubramani A, Lee YK, Whitley SK, Weaver BT, Shibata Y, Crawford GE, Hatton RD, Weaver CT. Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage. Immunity. 2010;32:616–27. doi: 10.1016/j.immuni.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shi G, Cox CA, Vistica BP, Tan C, Wawrousek EF, Gery I. Phenotype switching by inflammation-inducing polarized Th17 cells, but not by Th1 cells. J Immunol. 2008;181:7205–13. doi: 10.4049/jimmunol.181.10.7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lee YK, Turner H, Maynard CL, Oliver JR, Chen D, Elson CO, Weaver CT. Late developmental plasticity in the T helper 17 lineage. Immunity. 2009;30:92–107. doi: 10.1016/j.immuni.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Laurence A, Tato CM, Davidson TS, Kanno Y, Chen Z, Yao Z, Blank RB, Meylan F, Siegel R, Hennighausen L, Shevach EM, O’Shea JJ. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26:371–81. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 111.Pandiyan P, Conti HR, Zheng L, Peterson AC, Mathern DR, Hernandez-Santos N, Edgerton M, Gaffen SL, Lenardo MJ. CD4(+)CD25(+)Foxp3(+) regulatory T cells promote Th17 cells in vitro and enhance host resistance in mouse Candida albicans Th17 cell infection model. Immunity. 2011;34:422–34. doi: 10.1016/j.immuni.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen Y, Haines CJ, Gutcher I, Hochweller K, Blumenschein WM, McClanahan T, Hammerling G, Li MO, Cua DJ, McGeachy MJ. Foxp3(+) regulatory T cells promote T helper 17 cell development in vivo through regulation of interleukin-2. Immunity. 2011;34:409–21. doi: 10.1016/j.immuni.2011.02.011. [DOI] [PubMed] [Google Scholar]
- 113.Yang XP, Ghoreschi K, Steward-Tharp SM, Rodriguez-Canales J, Zhu J, Grainger JR, Hirahara K, Sun HW, Wei L, Vahedi G, Kanno Y, O’Shea JJ, Laurence A. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nature Immunology. 2011;12:247–54. doi: 10.1038/ni.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Das J, Ren G, Zhang L, Roberts AI, Zhao X, Bothwell AL, Van Kaer L, Shi Y, Das G. Transforming growth factor beta is dispensable for the molecular orchestration of Th17 cell differentiation. J Exp Med. 2009;206:2407–16. doi: 10.1084/jem.20082286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ghoreschi K, Laurence A, Yang XP, Tato CM, McGeachy MJ, Konkel JE, Ramos HL, Wei L, Davidson TS, Bouladoux N, Grainger JR, Chen Q, Kanno Y, Watford WT, Sun HW, Eberl G, Shevach EM, Belkaid Y, Cua DJ, Chen W, O’Shea JJ. Generation of pathogenic T(H)17 cells in the absence of TGF-beta signalling. Nature. 2010;467:967–71. doi: 10.1038/nature09447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–39. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Natoli G. Maintaining Cell Identity through Global Control of Genomic Organization. Immunity. 2010;33:12–24. doi: 10.1016/j.immuni.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 118.Birney E, Stamatoyannopoulos JA, Dutta A, Guig√≥ R, Gingeras TR, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Heintzman ND, Stuart RK, Hon G, Fu YT, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu CX, Ching KA, Wang W, Weng ZP, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature Genetics. 2007;39:311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 120.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang XM, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–12. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Bristow J, Ren B, Black BL, Rubin EM, Visel A, Pennacchio LA. ChIP-Seq identification of weakly conserved heart enhancers. Nature genetics. 2010;42:806–10. doi: 10.1038/ng.650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, Ragoussis J, Natoli G. Identification and Characterization of Enhancers Controlling the Inflammatory Gene Expression Program in Macrophages. Immunity. 2010;32:317–28. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 123.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–8. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21931–6. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2010 doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, Ku M, Durham T, Kellis M, Bernstein BE. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011 doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, Cobb BS, Yokomori K, Dillon N, Aragon L, Fisher AG, Merkenschiager M. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–33. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 128.Phillips JE, Corces VG. CTCF: Master Weaver of the Genome. Cell. 2009;137:1194–211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–8. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 130.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–7. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Navarro F, Lieberman J. Small RNAs guide hematopoietic cell differentiation and function. J Immunol. 2010;184:5939–47. doi: 10.4049/jimmunol.0902567. [DOI] [PubMed] [Google Scholar]
- 132.O’Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10:111–22. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 133.Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 134.Chong MM, Rasmussen JP, Rudensky AY, Littman DR. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J Exp Med. 2008;205:2005–17. doi: 10.1084/jem.20081219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liston A, Lu LF, O’Carroll D, Tarakhovsky A, Rudensky AY. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J Exp Med. 2008;205:1993–2004. doi: 10.1084/jem.20081062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhou X, Jeker LT, Fife BT, Zhu S, Anderson MS, McManus MT, Bluestone JA. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med. 2008;205:1983–91. doi: 10.1084/jem.20080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kuchen S, Resch W, Yamane A, Kuo N, Li Z, Chakraborty T, Wei L, Laurence A, Yasuda T, Peng S, Hu-Li J, Lu K, Dubois W, Kitamura Y, Charles N, Sun HW, Muljo S, Schwartzberg PL, Paul WE, O’Shea J, Rajewsky K, Casellas R. Regulation of MicroRNA expression and abundance during lymphopoiesis. Immunity. 32:828–39. doi: 10.1016/j.immuni.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rossi RL, Rossetti G, Wenandy L, Curti S, Ripamonti A, Bonnal RJ, Birolo RS, Moro M, Crosti MC, Gruarin P, Maglie S, Marabita F, Mascheroni D, Parente V, Comelli M, Trabucchi E, De Francesco R, Geginat J, Abrignani S, Pagani M. Distinct microRNA signatures in human lymphocyte subsets and enforcement of the naive state in CD4+ T cells by the microRNA miR-125b. Nat Immunol. 2011;12:796–803. doi: 10.1038/ni.2057. [DOI] [PubMed] [Google Scholar]
- 139.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–14. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Basso K, Sumazin P, Morozov P, Schneider C, Maute RL, Kitagawa Y, Mandelbaum J, Haddad J, Jr., Chen CZ, Califano A, Dalla-Favera R. Identification of the human mature B cell miRNome. Immunity. 2009;30:744–52. doi: 10.1016/j.immuni.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Stittrich AB, Haftmann C, Sgouroudis E, Kuhl AA, Hegazy AN, Panse I, Riedel R, Flossdorf M, Dong J, Fuhrmann F, Heinz GA, Fang Z, Li N, Bissels U, Hatam F, Jahn A, Hammoud B, Matz M, Schulze FM, Baumgrass R, Bosio A, Mollenkopf HJ, Grun J, Thiel A, Chen W, Hofer T, Loddenkemper C, Lohning M, Chang HD, Rajewsky N, Radbruch A, Mashreghi MF. The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat Immunol. 2010;11:1057–62. doi: 10.1038/ni.1945. [DOI] [PubMed] [Google Scholar]
- 142.Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S, Li Z, Wu Z, Pei G. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol. 2009;10:1252–9. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- 143.Lu LF, Boldin MP, Chaudhry A, Lin LL, Taganov KD, Hanada T, Yoshimura A, Baltimore D, Rudensky AY. Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell. 2010;142:914–29. doi: 10.1016/j.cell.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Han J, Kim D, Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc Natl Acad Sci U S A. 2007;104:12422–7. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, Kong B, Langerod A, Borresen-Dale AL, Kim SK, van de Vijver M, Sukumar S, Whitfield ML, Kellis M, Xiong Y, Wong DJ, Chang HY. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–9. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–63. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 147.Kapranov P, St Laurent G, Raz T, Ozsolak F, Reynolds CP, Sorensen PH, Reaman G, Milos P, Arceci RJ, Thompson JF, Triche TJ. The majority of total nuclear-encoded non-ribosomal RNA in a human cell is ‘dark matter’ un-annotated RNA. BMC Biol. 2010;8:149. doi: 10.1186/1741-7007-8-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, Taft RJ, Rinn JL, Ponting CP, Stadler PF, Morris KV, Morillon A, Rozowsky JS, Gerstein MB, Wahlestedt C, Hayashizaki Y, Carninci P, Gingeras TR, Mattick JS. The reality of pervasive transcription. PLoS Biol. 2011;9:e1000625. doi: 10.1371/journal.pbio.1000625. discussion e1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bernstein E, Allis CD. RNA meets chromatin. Genes and Development. 2005;19:1635–55. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- 150.Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–93. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–4. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Morales D Rivea, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–3. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 154.Sharma S, Findlay GM, Bandukwala HS, Oberdoerffer S, Baust B, Li Z, Schmidt V, Hogan PG, Sacks DB, Rao A. Dephosphorylation of the nuclear factor of activated T cells (NFAT) transcription factor is regulated by an RNA-protein scaffold complex. Proc Natl Acad Sci U S A. 2011;108:11381–6. doi: 10.1073/pnas.1019711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mourtada-Maarabouni M, Hedge VL, Kirkham L, Farzaneh F, Williams GT. Growth arrest in human T-cells is controlled by the non-coding RNA growth-arrest-specific transcript 5 (GAS5) J Cell Sci. 2008;121:939–46. doi: 10.1242/jcs.024646. [DOI] [PubMed] [Google Scholar]
- 156.Corcoran AE. The epigenetic role of non-coding RNA transcription and nuclear organization in immunoglobulin repertoire generation. Semin Immunol. 2010;22:353–61. doi: 10.1016/j.smim.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 157.Abarrategui I, Krangel MS. Regulation of T cell receptor-alpha gene recombination by transcription. Nat Immunol. 2006;7:1109–15. doi: 10.1038/ni1379. [DOI] [PubMed] [Google Scholar]
- 158.Richardson B. Primer: Epigenetics of autoimmunity. Nature Clinical Practice Rheumatology. 2007;3:521–7. doi: 10.1038/ncprheum0573. [DOI] [PubMed] [Google Scholar]
- 159.Patel DR, Richardson BC. Epigenetic mechanisms in lupus. Curr Opin Rheumatol. 2010;22:478–82. doi: 10.1097/BOR.0b013e32833ae915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ho SM. Environmental epigenetics of asthma: an update. J Allergy Clin Immunol. 2010;126:453–65. doi: 10.1016/j.jaci.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]