Abstract
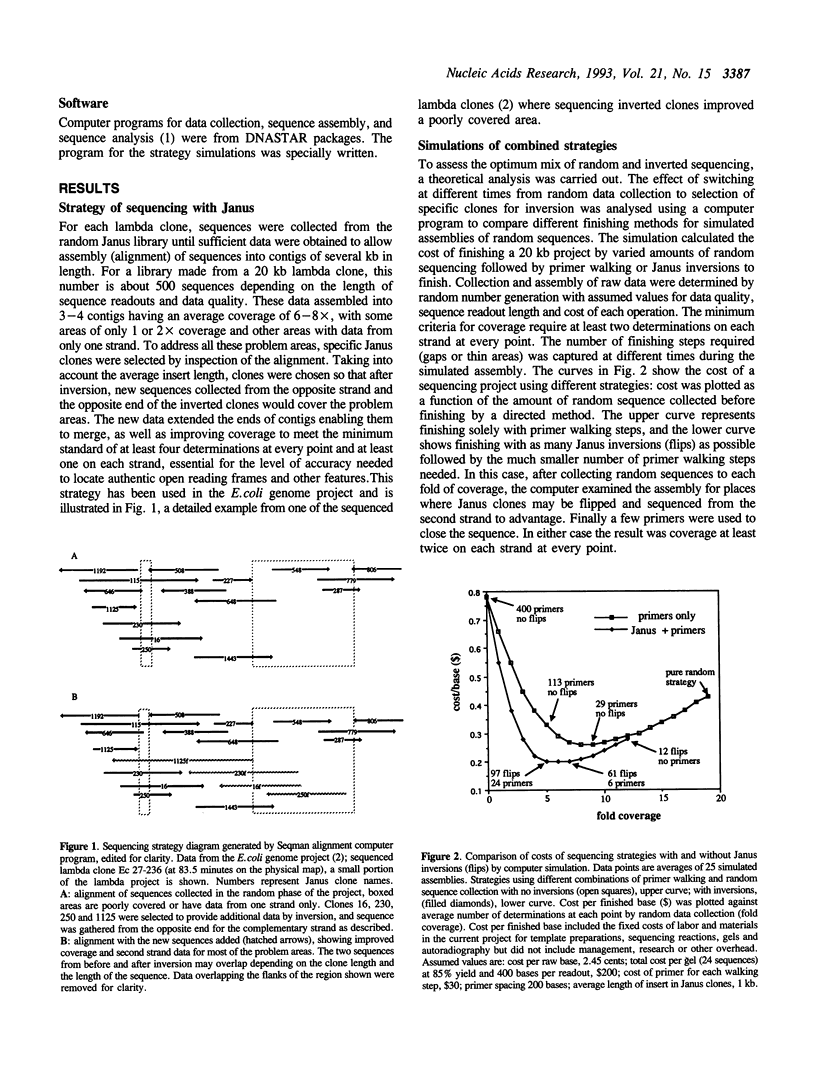
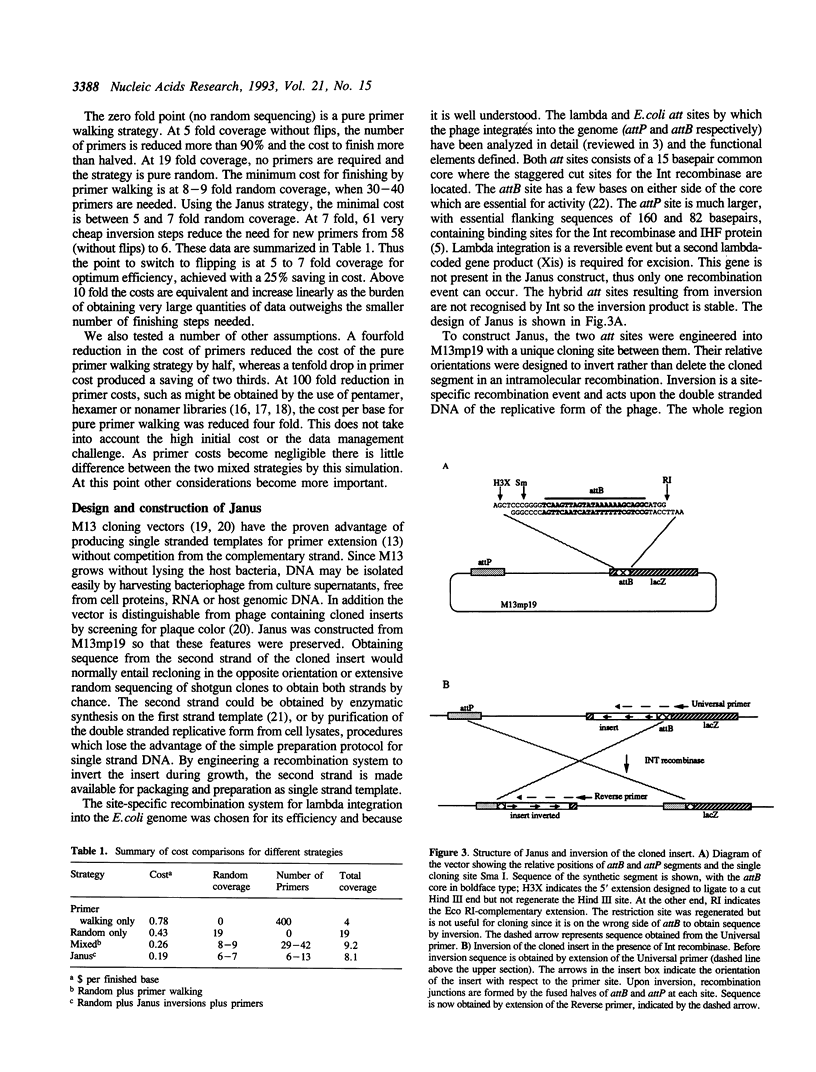
The design of large scale DNA sequencing projects such as genome analysis demands a new approach to sequencing strategy, since neither a purely random nor a purely directed method is satisfactory. We have developed a strategy that combines these two methods in a way that preserves the advantages of both while avoiding their particular limitations. Computer simulations showed that a specific balance of random and directed sequencing was required for the most efficient strategy, termed the Janus strategy, which has been used in the Escherichia coli genome sequencing project. This approach depended on obtaining sequence easily from either strand of a cloned insert, and was facilitated by inversion of the insert in the engineered M13 vector Janus, by site-specific recombination. The inversion was accomplished simply by growth on the appropriate host strain, when the DNA strand incorporated into the new single stranded phage was complementary to that in the original phage, and was sequenced by the same simple protocol as the first strand.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adachi T., Mizuuchi M., Robinson E. A., Appella E., O'Dea M. H., Gellert M., Mizuuchi K. DNA sequence of the E. coli gyrB gene: application of a new sequencing strategy. Nucleic Acids Res. 1987 Jan 26;15(2):771–784. doi: 10.1093/nar/15.2.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankier A. T., Weston K. M., Barrell B. G. Random cloning and sequencing by the M13/dideoxynucleotide chain termination method. Methods Enzymol. 1987;155:51–93. doi: 10.1016/0076-6879(87)55009-1. [DOI] [PubMed] [Google Scholar]
- Barnes W. M., Bevan M., Son P. H. Kilo-sequencing: creation of an ordered nest of asymmetric deletions across a large target sequence carried on phage M13. Methods Enzymol. 1983;101:98–122. doi: 10.1016/0076-6879(83)01008-3. [DOI] [PubMed] [Google Scholar]
- Burland V., Plunkett G., 3rd, Daniels D. L., Blattner F. R. DNA sequence and analysis of 136 kilobases of the Escherichia coli genome: organizational symmetry around the origin of replication. Genomics. 1993 Jun;16(3):551–561. doi: 10.1006/geno.1993.1230. [DOI] [PubMed] [Google Scholar]
- Daniels D. L., Plunkett G., 3rd, Burland V., Blattner F. R. Analysis of the Escherichia coli genome: DNA sequence of the region from 84.5 to 86.5 minutes. Science. 1992 Aug 7;257(5071):771–778. doi: 10.1126/science.1379743. [DOI] [PubMed] [Google Scholar]
- Dunn I. S., Blattner F. R. Charons 36 to 40: multi enzyme, high capacity, recombination deficient replacement vectors with polylinkers and polystuffers. Nucleic Acids Res. 1987 Mar 25;15(6):2677–2698. doi: 10.1093/nar/15.6.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. L., Ross W., Landy A. The lambda phage att site: functional limits and interaction with Int protein. Nature. 1980 May 8;285(5760):85–91. doi: 10.1038/285085a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innis M. A., Myambo K. B., Gelfand D. H., Brow M. A. DNA sequencing with Thermus aquaticus DNA polymerase and direct sequencing of polymerase chain reaction-amplified DNA. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9436–9440. doi: 10.1073/pnas.85.24.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieleczawa J., Dunn J. J., Studier F. W. DNA sequencing by primer walking with strings of contiguous hexamers. Science. 1992 Dec 11;258(5089):1787–1791. doi: 10.1126/science.1465615. [DOI] [PubMed] [Google Scholar]
- Kotler L. E., Zevin-Sonkin D., Sobolev I. A., Beskin A. D., Ulanovsky L. E. DNA sequencing: modular primers assembled from a library of hexamers or pentamers. Proc Natl Acad Sci U S A. 1993 May 1;90(9):4241–4245. doi: 10.1073/pnas.90.9.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landy A. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu Rev Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- Lee E. C., Gumport R. I., Gardner J. F. Genetic analysis of bacteriophage lambda integrase interactions with arm-type attachment site sequences. J Bacteriol. 1990 Mar;172(3):1529–1538. doi: 10.1128/jb.172.3.1529-1538.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Whalen W., Das A., Berg C. M. Rapid sequencing of cloned DNA using a transposon for bidirectional priming: sequence of the Escherichia coli K-12 avtA gene. Nucleic Acids Res. 1987 Nov 25;15(22):9461–9469. doi: 10.1093/nar/15.22.9461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Gronenborn B., Müller-Hill B., Hans Hopschneider P. Filamentous coliphage M13 as a cloning vehicle: insertion of a HindII fragment of the lac regulatory region in M13 replicative form in vitro. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3642–3646. doi: 10.1073/pnas.74.9.3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi M., Mizuuchi K. The extent of DNA sequence required for a functional bacterial attachment site of phage lambda. Nucleic Acids Res. 1985 Feb 25;13(4):1193–1208. doi: 10.1093/nar/13.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara O., Dorit R. L., Gilbert W. Direct genomic sequencing of bacterial DNA: the pyruvate kinase I gene of Escherichia coli. Proc Natl Acad Sci U S A. 1989 Sep;86(18):6883–6887. doi: 10.1073/pnas.86.18.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyamala V., Ames G. F. Genome walking by single-specific-primer polymerase chain reaction: SSP-PCR. Gene. 1989 Dec 7;84(1):1–8. doi: 10.1016/0378-1119(89)90132-7. [DOI] [PubMed] [Google Scholar]
- Siemieniak D. R., Slightom J. L. A library of 3342 useful nonamer primers for genome sequencing. Gene. 1990 Nov 30;96(1):121–124. doi: 10.1016/0378-1119(90)90350-z. [DOI] [PubMed] [Google Scholar]
- Smith V., Chee M. A simple method for sequencing the complementary strand of ssDNA from M13 clones. Nucleic Acids Res. 1991 Dec 25;19(24):6957–6957. doi: 10.1093/nar/19.24.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss E. C., Kobori J. A., Siu G., Hood L. E. Specific-primer-directed DNA sequencing. Anal Biochem. 1986 Apr;154(1):353–360. doi: 10.1016/0003-2697(86)90536-1. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]


