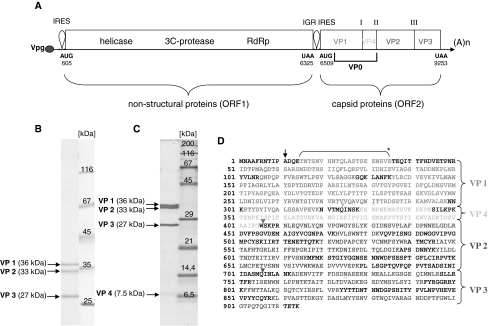
Fig. 1.
Genomic mapping of ABPV capsid proteins. (A) Schematic diagram of the genome of ABPV, which is classified as a dicistrovirus. The numbers represent nucleotide positions according to Govan et al. [25]. ORF1 encodes a helicase, a 3C-protease and an RNA-dependent-RNA polymerase (RdRp), whereas ORF2 encodes the four capsid proteins. The intergenic region (IGR) between the two ORFs acts as an internal ribosomal entry site (IRES). Roman numbers indicate processing sites. A purified ABPV suspension was separated in a 10% (B) or 15% PAA/0.1% SDS gel (C). The gel was stained with Coomassie blue G250. Arrows specify the viral capsid proteins VP1 to VP4. (D) Capsid proteins were excised from the gel, and their peptide composition was determined by MS/MS analysis after in-gel digestion with trypsin. The bracket (*) denotes a peptide fragment derived after cleavage of VP1 with protease V8. The black arrow points to the presumed translation initiation site [39]. Triangles signify putative cleavage sites within the capsid precursor [18, 25]. The amino acid sequence of the capsid precursor protein shown in (D) is almost identical to the ABPV strain “Germany 2” (gi|19068035) described by Bakonyi et al. [7]. Underlined amino acids indicate variable positions (colour figure online)