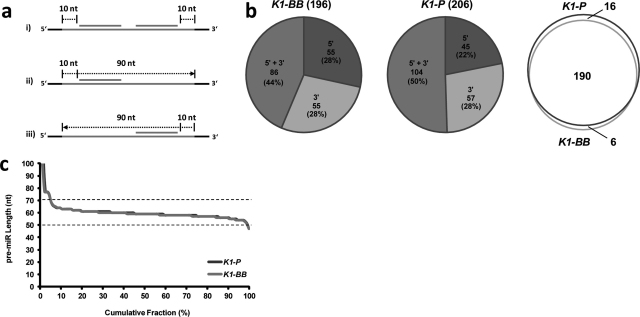
Fig. 3.
Characterization of CHO pre-miRNA sequences. (a) Scheme representing the strategy for pre-miRNA sequence extraction from a genomic locus mapped by either one or two mature miRNAs: (i) a buffer of 10 bases up- and downstream the mature miRNAs was taken in case both hairpin-arms were mapped. (ii) and (iii) For genomic positions aligned by a single miRNA a total pre-miRNA of 100 bases was extracted, starting 10 bases upstream a 5′ miRNA match or 10 bases downstream a 3′ match. (b) Distribution of CHO miRNA loci identified by alignment of either 5′ or 3′ mature miRNAs or both is shown. Venn overlap of miRNA genomic loci as identified independently in each CHO-K1 genomic reference sequence. (c) For pre-miRNA genomic loci mapped at both the 5′ and 3′ miRNA hairpin-arm, length of the pre-miRNA was calculated as the distance between the start of the 5′ miRNA alignment and the end of the 3′ miRNA alignment. Cumulative fraction of pre-miRNAs is plotted against the pre-miRNA length, showing that for most pre-miRs length ranged between 50 and 70 bases.