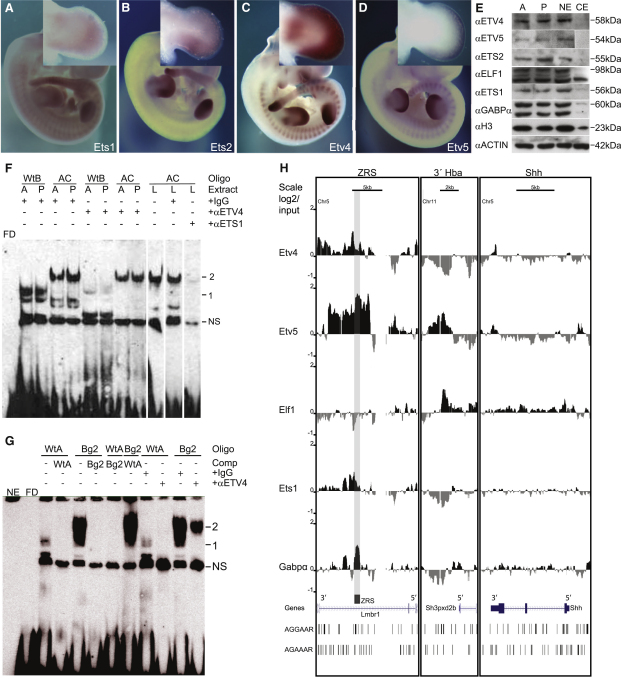
Figure 2.
ETS Factors Are Expressed in the Limb and Bind to the ZRS
(A–D) Whole-mount in situ hybridization analysis for Ets1 (A), Ets2 (B), Etv4 (C), and Etv5 (D) are shown in E11.5 embryos and limb buds.
(E) Western blot analysis using antibodies raised against ETS factors, designated αETV4, αETV5, αETS2, αELF1, αETS1, and αGABPα, and against histone H3 (αH3) and actin (αactin), with nuclear extracts from the anterior (A) and posterior (P) halves of the limb buds (E11.5). Also shown is a comparison between limb nuclear extracts (NE) and cytoplasmic extracts (CE). αH3 and αactin were used as loading controls.
(F) EMSA shows WtB and AC ds-oligo binding in nuclear extracts depleted for ETV4 or ETS1 using specific antibodies (αEtv4 and αEts1). (IgG was used as a nonspecific control.) Extracts from anterior (A) or posterior (P) halves or whole limbs (L) from E11.5 limb buds were used. Band 1 observed with the WtB probe was specifically depleted by the addition of αETV4 antibody, while Band shift 2 observed with the AC probe was specifically depleted by the addition of αETS1 antibody.
(G) EMSAs were conducted with ds-oligos containing the sequence for the wild-type ETV4 site A (WtA) or the Belg2 mutation (Bg). WtA ds-oligo shows a specific band (1) while that for Bg sequence gives an additional higher migrating band (2). The anti-ETV4 antibody depletes Band 1 observed with WtA and Bg probes (nonspecific IgG used as control).
(H) ChIP using antibodies to five different ETS factors (ETV4, ETV5, Elf1, ETS1, and GABPα) analyzed by hybridizing to tiling microarrays. Summary is presented using three different genomic regions, the y axis is Log2 for each ChIP/input DNA and the x axis represents a segment of DNA from the microarray. The DNA region containing the ZRS is highlighted by the gray shading. As controls, the whole of the Shh coding region plus promoter (Shh) and the region downstream of the α-globin locus (3′Hba) are shown. Scale bars are shown at the top and the positions of potential ETS1/GABPα (AGGAAG/A) and ETV4/ETV5 (AGAAAG/A) binding sites at the bottom of each panel.