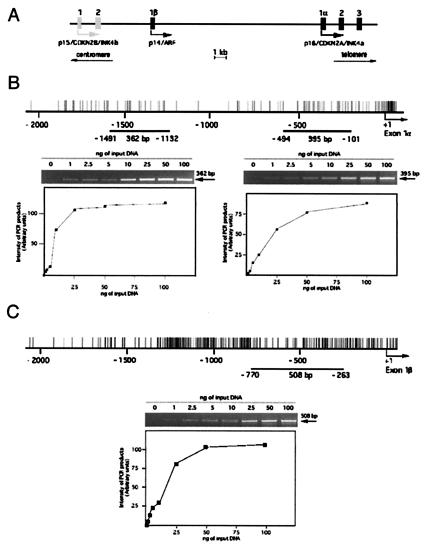
Figure 1.
Sensitivity of PCR amplification on sonicated DNA after DNA–protein cross-linking. (A) Schematic representation of the p15/p14/p16 locus on human chromosome 9p21. Shaded boxes, coding exons for p15/CDKN2B/INK4b (exons 1 and 2); solid boxes, coding exons for p14/ARF (exon 1β) and p16/CDKN2A/INK4a (exons 1α, 2, and 3); arrows, transcription initiation sites. The p16 and p14 promoters are separated by 20 kb and both are CpG sites. Distribution of CpG sites are represented by upward strokes for p16 (B) and p14 (C). Lines, regions amplified by PCR from the initiation of transcription of p16 (exon 1α) (B) or p14 (exon 1β) (C). Sensitivity of the PCR amplification was monitored for ChIP experiments in which the yield of PCR product depended on the amount of input DNA and 10% of the PCR mixture (total volume, 50 μl) from serial dilution of input DNA from HCT15 colon cells was analyzed on ethidium bromide-stained 2% agarose gels. The intensity of the bands of PCR products was measured by densitometry and plotted against the amount of input DNA (from 1 to 100 ng) for each set of primers. (B) The 362-bp band corresponds to the expected size of the PCR product for the Alu element (15 CpG sites, positions −1,491 to −1,132); the 395-bp band corresponds to the promoter of p16 (21 CpG sites, positions −494 to −101). (C) The 508-bp band corresponds to the expected size of the PCR product for the p14 promoter (43 CpG sites, positions −770 to −263).