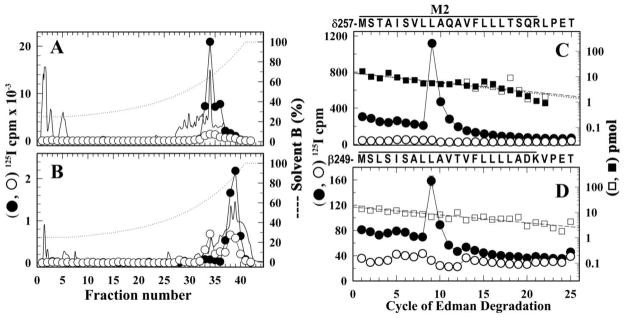
Figure 6. Identification of amino acids photolabeled by [125I]-SADU-3-72 in the δ M2 and βM2 segments in the Torpedo nAChR.
Affinity-purified nAChR was photolabeled with [125I]-SADU-3-72 in the presence of α-BgTx (●) or Carb (○) as detailed in Experimental Procedures. Shown are reversed-phase HPLC fractionation (A, B) and amino acid sequence analysis (C, D) of [125I]-SADU-3-72-labeled proteolytic fragments δT-5K (A, C) and βT-7K (B, D). The elution of peptides during HPLC was monitored by absorbance at 210 nm (solid line) and 125I elution was quantified by γ counting of each fraction (●, ○). C, 125I (●, ○) and PTH- amino acids (■, □) released during amino acid sequence analysis of 125I peak (fractions 33–35; ●, 20,000 cpm; ○, 2,000 cpm) from the HPLC purification of δT-5K. In each sample, the primary peptide detected began at δMet257 (●, I0 = 13.8 ± 2.4 pmol, R = 92%; ○, I0 = 14 ± 1.5 pmol, R = 92%) and for the (●) sample there was a peak of 125I cpm release in cycle 9 corresponding to labeling of δLeu265 (30 cpm/pmol). D, 125I (●, ○) and PTH-amino acids (■, □) released during amino acid sequence analysis of 125I peak (fractions 37–40; ●, 5,000 cpm; ○, 1,700 cpm) from the HPLC purification of βT-7K. In each sample, the primary peptide detected began at βMet249 (●, I0 = 9 ± 1 pmol, R = 92%; ○, I0 = 16 ± 3 pmol, R = 93%) and for the (●) sample there was a peak of 125I cpm release in cycle 9 corresponding to labeling of βLeu257 (5 cpm/pmol).