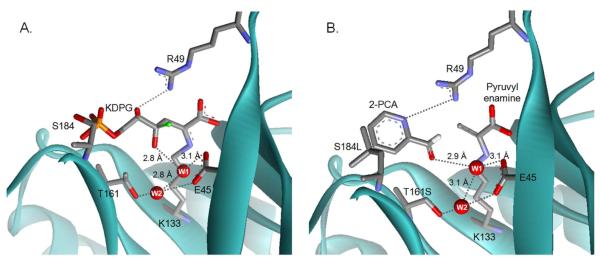
Figure 5.
Models of substrates bound to E. coli KDPG aldolase. (A) A model of KDPG bound to the active site of KDPG aldolase. A proposed hydrogen bond network of waters that stabilize the complex is shown in gray dashed lines with key distances indicated. Water W1 is proposed to preferentially enhance the affinity of the gluco-sugar and mediate catalytically important proton transfer steps. Water W2 is proposed to help position W1. The C4 hydroxyl from a model of a galacto-sugar bound to KDPG aldolase is superimposed as a green ball. This model suggests that the galacto-epimer is a less efficient substrate for KDPG aldolase because O4 bumps into R49, the angle for deprotonation of O4 by W1 is poor, and the distance between these atoms is long (3.8 Å). (B) A model of 2-PCA bound to the T161S/S184L mutant. The model suggests that hydrophobic contacts are made between the aromatic heterocycle of 2-PCA and leucine at position 184 and that the T161S mutation removes a steric clash with the aldehydic oxygen of 2-PCA. Also the side chain of R49 is positioned to form a hydrogen bond with N2 of 2-PCA, similar to the hydrogen bond between KDPG O4 and R49.