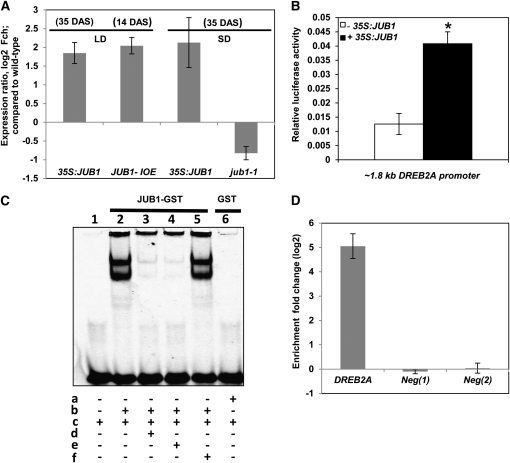
Figure 10.
DREB2A Is a Direct Target of JUB1.
(A) Expression of DREB2A in 35S:JUB1, JUB1-IOE, and jub1-1 lines compared with the wild type. Plant ages are indicated in days after sowing (DAS). LD, long day; SD, short day. Numbers on the y axis indicate expression fold change (log2 basis) compared with the wild type. Data represent means ± sd of five (LD) or three (SD) independent experiments.
(B) Transactivation of DREB2A expression (from its ~1.8-kb promoter) by JUB1 in Arabidopsis mesophyll cell protoplasts. The ProDREB2A:FLuc construct harboring the DREB2A promoter upstream of the firefly (Photinus pyralis) luciferase (FLuc) open reading frame was cotransformed with the 35S:JUB1 plasmid (omitted in control experiments). The 35S:RLuc vector was used for transformation efficiency normalization. Bars indicate the sd of at least four biological replicates. The asterisk indicates significant difference to control at P < 0.05.
(C) EMSA. Purified JUB1-GST protein binds specifically to the JUB1 binding site within the DREB2A promoter. In vitro DNA binding reactions were performed with the 40-bp wild-type fragment of the DREB2A promoter containing the JUB1 motif (5′-GATGCCGTTAGAGACACG-3′). a, GST protein; b, JUB1-GST protein; c, 5′-DY682 double-stranded oligonucleotide containing the perfect JUB1 binding site; d, 100× competitor (unlabeled oligonucleotide containing perfect JUB1 binding site); e, 200× competitor (unlabeled oligonucleotide containing perfect JUB1 binding site); f, 200× mutated oligonucleotide (unlabeled with mutation in JUB1 binding site where 5′-GATGCCGTTAGAGACACG-3′ was replaced by 5′-GATGCCAATAGAGACACG-3′).
(D) ChIP-qPCR. Whole shoots of 35-d-old Arabidopsis plants expressing GFP-tagged JUB1 under the control of the CaMV 35S promoter (35S:JUB1-GFP) and wild-type plants were harvested for the ChIP experiment. qPCR was used to quantify enrichment of the DREB2A promoter. As negative controls, primers annealing to promoter regions of two Arabidopsis genes lacking a JUB1 binding site, At3g18040 (Neg 1) and At2g22180 (Neg 2), were used. Data represent means ± sd of three independent experiments.