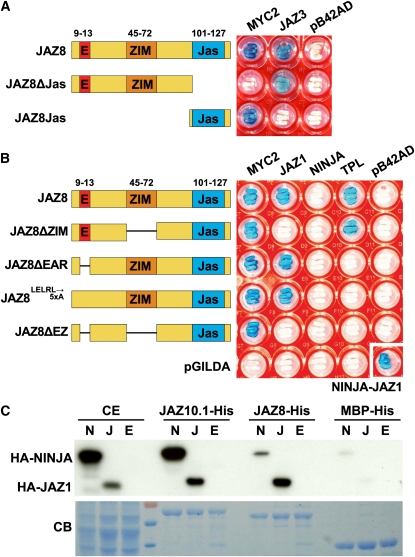
Figure 8.
Protein–Protein Interaction Domains in JAZ8.
(A) Y2H assay of JAZ8 and JAZ8 deletion derivatives with MYC2 and JAZ3. Yeast strains expressing both the bait (JAZ8) and prey (MYC2 or JAZ3) proteins were plated on media containing X-Gal. LacZ-mediated blue-color formation is indicative of protein–protein interaction. JAZ8 proteins were coexpressed with an empty prey vector (pB42AD) as a negative control. Photographic images of yeast cells were taken after 48 h of incubation at 30°C.
(B) Y2H analysis of JAZ8 deletion proteins with MYC2, JAZ1, NINJA, and TPL. Yeast strains expressing both the bait (JAZ8 or JAZ8 deletion) and prey (MYC2, JAZ1, NINJA, or TPL) proteins were tested as described in (A). Empty bait (pGILDA) and prey (pB42AD) vectors were used as negative controls. As a positive control for NINJA interaction, yeast cells were cotransformed with pB42AD-NINJA and pGILDA-JAZ1 (bottom right).
(C) JAZ8 interacts weakly with NINJA in vitro. Purified JAZ-His proteins (fused to MBP) were incubated with crude extract from yeast cells expressing HA-tagged derivatives of NINJA (N), JAZ1 (J), or an empty vector control (E). MBP-His was used as a control for specificity. Purified protein complexes were separated by SDS-PAGE and probed with an anti-HA antibody for the presence of HA-tagged NINJA (HA-NINJA) or JAZ1 (HA-JAZ1). The bottom panel shows a Coomassie blue (CB)–stained gel to visualize the amount of protein loaded. CE, crude yeast extracts that were used as an input control. Protein molecular weight markers were run in the fourth lane from the left.