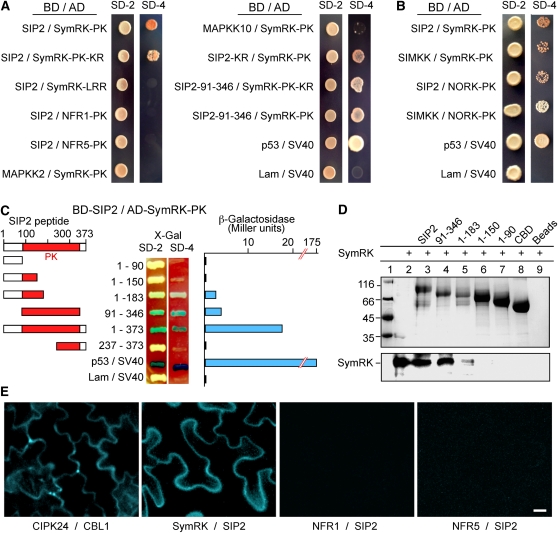
Figure 2.
Interaction of SIP2 with SymRK in Vitro and in Planta.
(A) SIP2 interacts with SymRK in yeast cells. Yeast Y187 cells carrying the Gal4 DNA binding domain (BD) fusion constructs were mated with yeast AH109 cells harboring the Gal4 activation domain (AD) fusion constructs. The diploid cells were selected on SD media lacking Leu and Trp (SD-2), and interaction was assessed according to their ability to grow on selective SD media lacking Leu, Trp, His, and Ade (SD-4) for 5 d. SymRK-LRR, the extracellular LRR region of SymRK; SymRK-PK, the intracellular PK domain of SymRK; NFR1-PK, the intracellular PK domain of NFR1; NFR5-PK, the intracellular PK domain of NFR5; SIP2-KR, a Lys-to-Arg substitution kinase-negative mutant of SIP2; SIP2-91-346, the PK domain of SIP2. Note that SIP2 did not interact with the putative NF receptors (NFR1 and NFR5), and SymRK did not interact with Lj-MAPKK2 and Lj-MKK10. SIP2 did interact with the SymRK ortholog (NORK) from alfalfa, while SymRK also interacted with the SIP2 ortholog (SIMKK) from alfalfa. The interaction between mammalian p53 and SV40 served as a positive control, whereas coexpression of lamin (Lam) and SV40 served as a negative control.
(B) Interactions between SymRK and SIP2 orthologs from Lotus and Medicago. Lotus SymRK interacted with the SIP2 ortholog, SIMKK, from alfalfa, and similarly, Lotus SIP2 interacted with the SymRK ortholog, NORK, from alfalfa. The interaction between alfalfa SIMKK and NORK-PK was relatively weak, and the yeast colonies were selected on SD/-Leu/-Trp/-Ade media, followed by selection on SD/-Leu/-Trp/-Ade/-His.
(C) The PK domain of SIP2 is responsible for interaction with SymRK. SIP2 and its truncated constructs were expressed as fusion proteins with the GAL4 binding domain (BD) in pGBKT7. SymRK-PK was expressed as a recombinant protein fused with the GAL4 activation domain (AD) in pGADT7. Yeast cells containing both plasmids were grown on SD-Leu-Trp medium (SD-2) containing X-Gal (80 mg/L) and assessed for interactions on SD-Leu-Trp-His-Ade medium (SD-4) containing X-Gal. The strength of interaction was evaluated by assaying β-galactosidase activities in soluble extracts prepared from yeast colonies.
(D) In vitro protein pull-down assay for the interaction between SIP2 and SymRK-PK. CBD-tagged SIP2 and four of its truncated products (numbers indicate residues of SIP2) were absorbed to chitin beads and mixed with purified soluble SymRK-PK. After washing, proteins pulled down by the chitin beads were separated via SDS-PAGE and visualized with Coomassie blue dye (top). The same gel was used for immunoblotting with anti-SymRK antibody (bottom). Note that the full-length SIP2 and its PK domain (residues 91 to 346) could pull down SymRK, and the N-terminal half (1 to 183) of SIP2 also could interact with SymRK with a reduced affinity.
(E) BiFC on the interaction between SIP2 and SymRK in planta. N. benthamiana leaves were cotransformed with SCFPC155:SymRK-full and SIP2-full:SCFPN173 (SymRK / SIP2). Leaf epidermal cells were observed via fluorescence microscopy. Because the CFP is split into N- and C-terminal halves and fused with different proteins, no cyan fluorescence would be observed if the fusion proteins did not interact. As a positive control, Arabidopsis CIPK24 (CBL-interacting protein kinase 24) and calcineurin B-like 1 (CBL1) were fused with the C-terminal CFPC155 and N-terminal CFPN173, respectively, and coexpressed. Note that no interactions were observed between NFR1 and SIP2, or NFR5 and SIP2 using similar fusion approach. Bar = 20 μm.