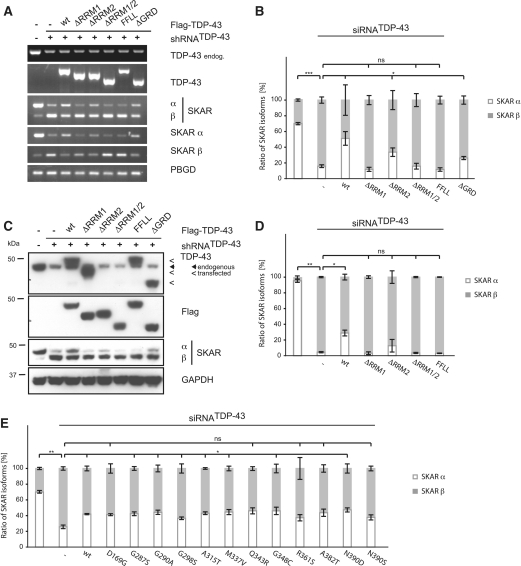
Figure 3.
SKAR alternative splicing is dependent on RRM1 of TDP-43. (A) Stably silenced HEK293E cells (shRNATDP-43) or transiently silenced HEK293 cells (siRNATDP-43) were transiently transfected with either control vector (−) or Flag-TDP-43 variants (wt, ΔRRM1, ΔRRM2, ΔRRM1/2, FFLL and ΔGRD or disease-associated mutations, as indicated). Parental HEK293E cells or cells treated with a scrambled siRNA (−) were used as an internal control. (A) Total RNA was extracted and subjected to semi-quantitative RT–PCR using primer pairs amplifying total TDP-43, endogenous TDP-43, total SKAR (ex2–ex4), SKAR α (ex2|3–ex4), SKAR β (ex2|4–ex4) and PBGD as a housekeeping gene. (B and E) RNA was extracted and real-time PCR performed with primer pairs against SKAR α (ex2|3–ex4) (white bars), SKAR β (ex2|4–ex4) (gray bars) and total SKAR (ex5|6–ex7). PBGD was used as a housekeeping gene. Resulting relative SKAR α/PBGD, SKAR β/PBGD and total SKAR/PBGD ratios were re-calculated into absolute copy values and normalized to total SKAR values. Original qRT data is presented in Supplementary Figure S1C and S1D, respectively. *P < 0.05; **P < 0.005; ***P < 0.0005; ns = not significant. (C and D) Protein was extracted, electrophoresed and resulting western blots probed with anti-TDP-43, anti-Flag and anti-SKAR antibodies. GAPDH was used as a loading control. (D) Shown are the mean values ± SEM of densitometric analysis of three independent experiments. *P < 0.05; **P < 0.005; ns = not significant.