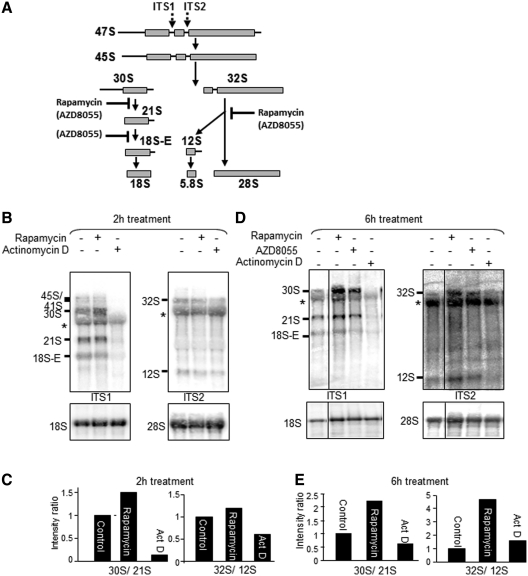
Figure 3.
Rapamycin affects pre-rRNA processing in HeLa cells. (A) Schematic diagram of pre-rRNA processing pathways in HeLa cells. Structure of the primary 47S rRNA transcript containing two internal transcriber spacers (ITS1 and ITS2) and two external transcribed spacers. The 47S pre-rRNA is processed through intermediate precursors to mature 18S, 28S and 5.8S rRNAs. The locations of the probes used are shown (ITS1 and ITS2). (B and D) effects of rapamycin and AZD8055 (each used at 100 nM) on rRNA processing in serum-fed cells. Northern hybridization of total RNA from HeLa cells after treatment with rapamycin or actinomycin D (20 ng/ml). After 2–6 h treatment (as indicated), total RNA was extracted and subjected to northern blotting analysis for pre-rRNA species. Membranes were hybridized with the ITS1 and ITS2 probes and probes for 18S and 28S rRNAs. The asterisk indicates cross-reaction with 28S rRNA and the vertical line denotes that these are non-adjacent lanes from the same gel/northern blot. (C and E) Ratios of different pre-rRNA species. Bar graphs show signal quantification using a phosphorimager and the bars indicate the ratios of 30S to 21S and 32S to 12S after rapamycin or actinomycin D treatment.