Abstract
Homing endonucleases (HEs) promote the evolutionary persistence of selfish DNA elements by catalyzing element lateral transfer into new host organisms. The high site specificity of this lateral transfer reaction, termed homing, reflects both the length (14–40 bp) and the limited tolerance of target or homing sites for base pair changes. In order to better understand molecular determinants of homing, we systematically determined the binding and cleavage properties of all single base pair variant target sites of the canonical LAGLIDADG homing endonucleases I-CreI and I-MsoI. These Chlorophyta algal HEs have very similar three-dimensional folds and recognize nearly identical 22 bp target sites, but use substantially different sets of DNA-protein contacts to mediate site-specific recognition and cleavage. The site specificity differences between I-CreI and I-MsoI suggest different evolutionary strategies for HE persistence. These differences also provide practical guidance in target site finding, and in the generation of HE variants with high site specificity and cleavage activity, to enable genome engineering applications.
INTRODUCTION
The lateral transfer of mobile introns or inteins into new host organisms is termed ‘homing’ (1–4). The homing reaction has three specific components: the laterally transferred genetic element, typically a self-splicing intron or intein; a homing endonuclease (HE) protein, often encoded within the mobile intron or intein; and a target or ‘homing’ site that can be cleaved by a cognate HE. Target site cleavage initiates recombination-mediated repair using the homologous intron or intein-containing donor allele as a repair template. Successful repair results in transfer of the mobile intron or intein into the cleaved host target site to create a new intron-containing (or ‘intron+’) allele. Intron/intein insertion disrupts the target site, and thus prevents additional rounds of cleavage that could result in intron loss. Homing is thus an efficient, unidirectional lateral transfer mechanism that ensures the potential for additional rounds of intron or intein lateral transfer into HE cleavage-sensitive hosts (4).
Several hundred HEs encoded by mobile introns or inteins have been identified across all domains of life. These HEs have been categorized into five families on the basis of shared sequence motifs: the LAGLIDADG, HNH, His-Cys box, GIY-YIG and PD-(D/E)XK HE families (5,6). The LAGLIDADG homing endonuclease (LHE) family, with several hundred members, is the largest and best-studied of these families (7,8). LHE proteins share an αββαβα core fold in which the conserved ‘LAGLIDADG’ protein motif forms a dimerization or intra- molecular folding interface, and contributes catalytic acidic aspartic or glutamic acid residues to the nuclease active sites. DNA target site recognition is mediated by contacts made by LHE amino acid side chains to DNA bases or to the phosphodiester backbone. Most of these interface amino acid residues are located in anti-parallel β sheets flanking the LAGLIDADG interface.
The high site specificity of LHE cleavage reflects the large number of phased, direct and water-mediated contacts made by LHEs to target site DNA (5). Despite their high site specificity, many LHEs appear to tolerate some target site base pair changes without a loss of site binding or cleavage [see, e.g. (9,10)]. This seemingly paradoxical combination—high site specificity with the ability to tolerate target site base pair changes—may be evolutionarily advantageous: high site specificity minimizes toxicity to current hosts by limiting off-target cleavage, while the ability to tolerate target site genetic variation may maximize the potential for continued lateral spread (11–14).
In order to better understand the molecular determinants of homing and lateral transfer, we determined the ability of two well-characterized, homologous LHE proteins to bind and to cleave all single base pair variants of their native DNA target sites. The proteins, I-CreI and I-MsoI, are encoded by mobile introns in the chloroplast DNAs of different Chlorophyta algal species. Both homodimeric endonucleases share a common three-dimensional fold and 22 bp target sites that differ at only 2 of 22 bp positions. The I-CreI and I-MsoI target sites are pseudo-palindromic, reflecting the homodimeric architecture of both endonucleases, and have left and right halves that share, respectively, 7 of 11 and 5 of 11 bases (Figure 1) (15,16). Our prior structural analyses demonstrated that I-CreI and I-MsoI use substantially different sets of DNA–protein contacts to recognize their target sites (Figure 1C) (17,18). In work reported here, we systematically determined the in vitro binding and cleavage properties of I-CreI, I-MsoI and the monomeric versions mCreI and mMsoI (19) on all single base pair variant DNA target sites (Figure 1). We also determined the in vivo cleavage specificity profiles for mCreI and mMsoI in human cells using the same target site libraries.
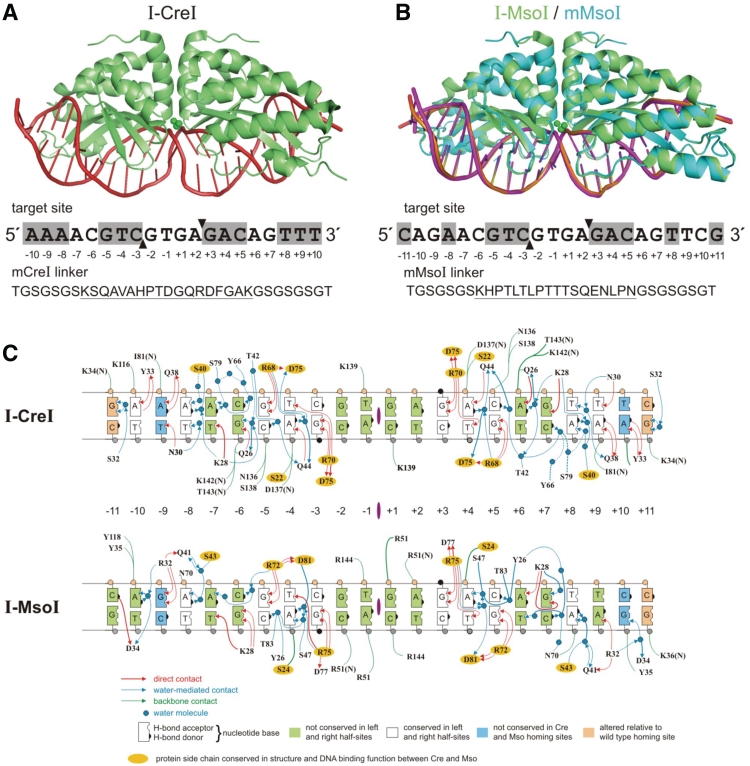
Figure 1.
I-CreI, I-MsoI and their monomeric variants. (A) Crystal structure of I-CreI (green) bound with target site DNA (red) (17). The I-CreI target site is shown below the co-crystal structure, with target site nucleotide positions numbered relative to the center of symmetry. Two-fold symmetric, palindromic target site positions are shown in shaded boxes, and the location of top and bottom strand cleavage sites by filled arrows. mCreI, a monomeric/single chain version of I-CreI, was generated by connecting the two I-CreI domains with a 33 amino acid residue linker whose sequence is shown below the target site in single letter amino acid code. The unique portion of the linker, located between flexible GS repeats, is underlined (19). (B) Crystal structures of I-MsoI (green) and mMsoI (cyan) bound with their DNA target site (18,19). The I-MsoI target site, location of palindromic target site positions and the location of cleavage sites are indicated as in (A) above. The 33 residue linker sequence used to create mMsoI is indicated below the target site in single letter amino acid code with the unique portion of the linker underlined (19). (C) DNA–protein interfaces of I-CreI and I-MsoI bound to native DNA target sites. The DNA–protein contact maps for I-CreI (top) and I-MsoI (bottom) were determined from their respective co-crystal structures (17,18) and redrawn in common format. Protein side chains that are conserved in structure and DNA binding function are indicated by yellow ovals. Base pairs at the ±11 positions, shown in pink, differed from native target site base pairs and were included in successful crystallization oligonucleotides.
MATERIALS AND METHODS
Materials
The bacterial protein expression plasmids pET15b and pET24d were obtained from Novagen (Gibbstown, NJ, USA). The Escherichia coli protein expression host strain C2566 was obtained from New England Biolabs (Ipswich, MA, USA). DNA oligonucleotides (50-nmol scale, salt-free) were synthesized by Operon (Huntsville, AL, USA). Qiaquick PCR purification kits and Ni-NTA HisSorb plates were obtained from Qiagen (Valencia, CA, USA). Other reagents, including restriction enzymes, Taq DNA polymerase and T4 DNA ligase were obtained from New England Biolabs (Ipswich, MA, USA) or Sigma-Aldrich (St Louis, MO, USA).
Protein expression and purification
Homing endonuclease proteins were expressed and purified as previously described by nickel affinity chromatography (19). Proteins for in vitro binding assays were expressed from pET15b with an N-terminal 6 × His affinity purification/binding tag. Proteins used for in vitro cleavage assays were expressed and purified from pET24d without affinity tags.
In vitro binding specificity
The in vitro target site binding affinities were determined using a competitive binding assay as previously described (20). In brief, proteins with N-terminal 6 × His tags were immobilized in Ni-NTA HisSorb 96-wells plates (Qiagen) by incubating 200 µl of 100 nM protein in TBS/BSA buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.2% BSA) with plate wells for 2 h at room temperature. Unbound protein was removed by washing plates four times with TBS/Tween-20 [50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.05% Tween-20]. Immobilized proteins were then incubated with a mixture of 100 nM fluorescently labeled native target site DNA and 3 µM (a 30-fold excess) of each of three unlabeled competitor target site DNAs containing alternative base pair substitutions at a given target site base pair position in 200 µl of binding buffer [50 Mm Tris, pH 7.5, 150 mM NaCl, 0.02 mg/ml poly(dI-dC), 10 mM CaCl2]. After incubation for 4 h at room temperature, plates were washed four times with TBS (50 mM Tris, pH 7.5, 150 mM NaCl). Retained fluorescence was quantified on a SpectraMax® M5/M5e microplate reader (Molecular Devices; excitation, 510 nm; emission, 565 nm; cutoff, 550 nm). All measurements were performed in triplicate, and relative in vitro binding affinities were calculated using the following formula:
where F(x), F(t) and F(n) indicate fluorescence intensities of wells containing immobilized protein that were incubated with unlabeled base substitution target sites [F(x)] or with the native [F(t)] or a scrambled sequence target site [F(n)].
In vitro cleavage specificity
I-CreI or I-MsoI target site were synthesized as pairs of complementary DNA oligonucleotides which were annealed and ligated into the XhoI and SacI sites of the recombination reporter plasmid pDR-GFP-univ, a universal target site version of pDR-GFP (19,21) (http://depts.washington.edu/monnatws/plasmids/pDR-GFP-univ.pdf). The I-CreI site library consisted of 61 unique target site plasmids: a native I-CreI target site plasmid and 60 additional plasmids with single base pair substitutions covering target site positions −10 to +10. The corresponding I-MsoI site library consisted of 67 unique target site plasmids: a native I-MsoI target site plasmid and 66 additional plasmids with single base substitutions covering target site positions −11 to +11. Each target site was amplified from pDR-GFP-univ plasmid DNA using pairs of primers that placed the target site at the center of different-sized amplicon products. Amplicons harboring target site A substitutions were 2200 bp long, and those with C, G or T substitutions were, respectively, 1900, 1600 or 1320 bp long (Figure 3A). After purification, equimolar amounts of the four DNA substrate fragments were combined to generate 20 I-CreI (or 22 I-MsoI) substrate pools for competitive in vitro cleavage assays.
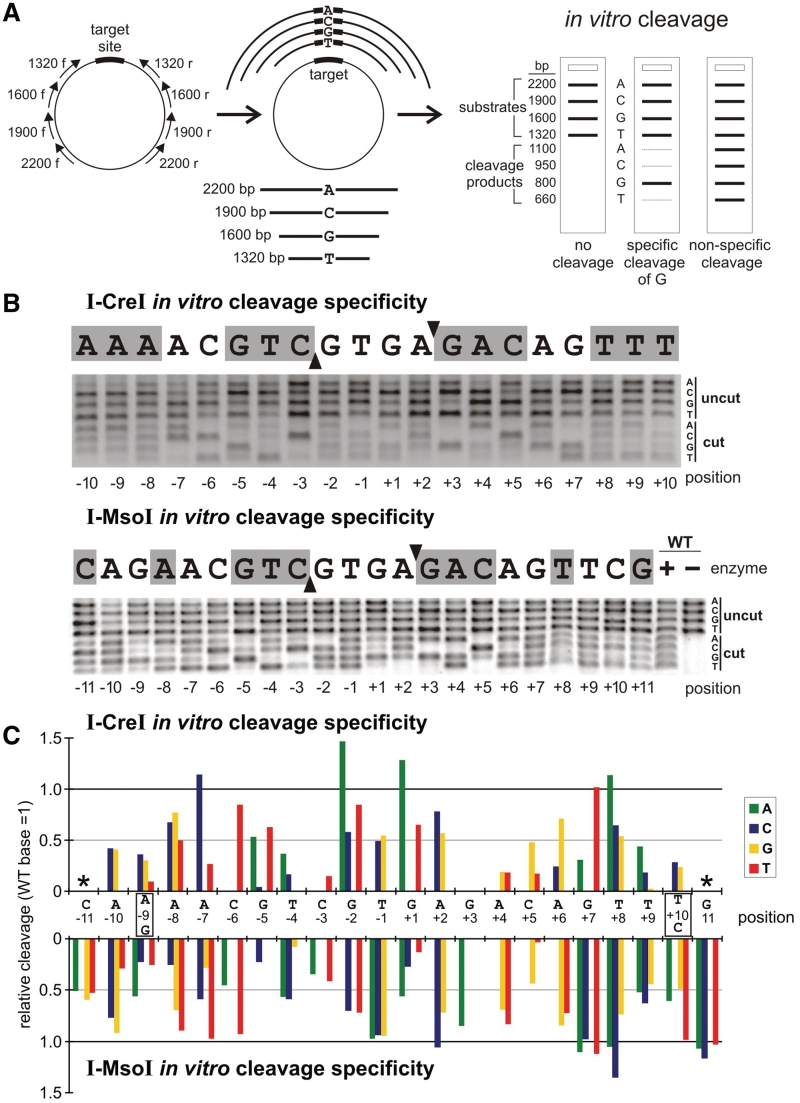
Figure 3.
In vitro cleavage specificity profiles of I-CreI and I-MsoI. (A) Outline of the ‘bar code’ cleavage assay used to assess the comparative cleavage efficiency of target sites with base pair substitutions (see Methods). Hypothetical base-specific or non-specific cleavage patterns are depicted at right (25). (B) Agarose gels displaying in vitro cleavage specificity profiles for I-CreI and I-MsoI determined as described in panel (A). (C) Quantitation of cleavage specificity data in (B), where bar heights indicate extent of cleavage of base pair substitutions relative to the native base pair whose cleavage efficiency has been arbitrarily set to 1.0. All results are the mean of three replicates in which the standard deviations were ±5%. Error bars have been omitted for graphical clarity.
HE proteins for in vitro cleavage assays were expressed in E. coli host strain C2566 from pET24d and purified as previously described (19). In vitro cleavage assays were conducted in 20 mM Tris pH 8.0, 10 mM MgCl2 with 1:1 protein/DNA ratio under conditions where ∼50% of the wild-type target site was cleaved. This corresponded to 15 min at 37°C for I-CreI/mCreI, and 1 h at 37°C for I-MsoI/mMsoI. Digests were stopped by adding loading buffer containing 0.1% SDS to samples, and the ladders of substrate and product fragments from each digest were separated by agarose gel electrophoresis. Fragment band intensities were quantified after staining using ImageQuant (Molecular Dynamics). Target site cleavage efficiency was calculated from the ratio of substrate to product bands, normalized to the cleavage efficiency of the native base pair at each target site position to generate a relative in vitro cleavage index.
In vitro competitive cleavage assay
The plasmid substrate for in vitro competitive cleavage assays were constructed by cloning both a native and a test target site into pCcdB (22) at two different locations: the native target site into AflIII/BglII-cleaved plasmid DNA, and the test target site into NheI/SacII-cleaved plasmid DNA. In order to perform competitive cleavage assays, plasmid substrates were linearized by XbaI digestion, and 100 ng of linear plasmid substrate was incubated with LHEs in the presence of 20 mM Tris pH 8.0, 100 mM NaCl, 10 mM MgCl2 for 1 h at 37°C. Cleavage reactions were quenched by adding 10 mM EDTA and 1% SDS followed by heating for 10 min at 60°C. Plasmid substrate and cleavage products were separated by agarose gel electrophoresis, visualized by staining with ethidium bromide and photographed for quantitation.
In vivo cleavage/recombination assays
The ability of mCreI and mMsoI to cleave target sites in human cells was determined using a human cell transient transfection assay as previously described [Figure 4A; (19)]. In brief, 293 T cells (3 × 105 cells/well in 500 µl of growth medium) were plated in 24-well plates 1 d prior to transfection and grown in a 37°C humidified, 5% CO2 incubator. Complete growth medium consisted of Dulbecco-modified Eagle's medium (Cellgro) supplemented with 10% fetal bovine serum (Cellgro) and 1% penicillin/streptomycin (Gibco). Wells were 50–80% confluent at the time of co-transfection with a pDR-GFP-univ target site plasmid and a coding plasmid for either mCreI or mMsoI. Transfections contained a total of 1.5 µg plasmid DNA/well (3:1 molar ratio of expression to target site plasmid DNA), and were performed overnight at 3% CO2 using a modified calcium phosphate protocol (19,23).
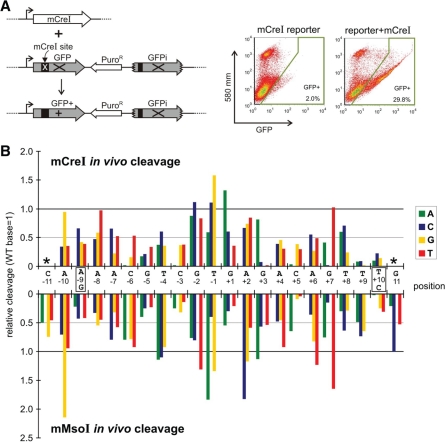
Figure 4.
The in vivo cleavage specificity profiles of mCreI and mMsoI. (A) Assay used to measure in vivo cleavage efficiency of variant target sites in human cells, where target site cleavage leads to the generation of GFP+ recombinant cells. (B) In vivo cleavage specificity profiles of mCreI and mMsoI, determined as described in A by co-transfecting endonuclease coding and target site plasmids into human 293T cells. Relative cleavage efficiencies are plotted as in Figure 3 with the native base pair GFP+ value arbitrarily set to 1.0. All assay values represent the mean of three replications, with error bars omitted for graphical clarity.
Transfected cells were analyzed by flow cytometry 48 h after transfection to quantify the frequency of generation of cleavage-depenent GFP+ recombinant cells. In brief, cells were tryspinized and washed in PBS, resuspended at ∼106 cells/ml in PBS buffer and then stained with propidium iodine (10 ng/µl) prior to analysis on an Influx flow cytometer (Cytopeia). Typically 50 000 events were scored for each transfected sample by gating log side versus linear forward scatter and for PI exclusion to quantify the fraction of GFP+ viable cells. Transfection efficiency was monitored in all experiments by using a GFP+ positive control vector pEGFP-C1 (Clontech) in the same experiment. In vivo cleavage efficiencies for single base pair variant target sites were calculated from the percent GFP+ cells, corrected for transfection efficiency and normalized against the GFP+ frequency of the native base pair at each target site position.
RESULTS
In vitro binding specificity
Target site binding affinities for all four proteins were determined using a competitive binding assay (20). In brief, N-terminal 6 × His-tagged HE proteins were immobilized in 96-well plates, then incubated with a fluorescently labeled native target site oligonucleotide followed by a 30-fold molar excess of unlabeled competitor target site DNA. Competitor sites included the native target site, single base pair variant target sites or an unrelated sequence as a control for non-specific binding. Relative binding affinities were calculated from competitor concentration-specific loss of fluorescence. Figure 2 displays the in vitro binding profiles for the native homodimers I-CreI and I-MsoI.
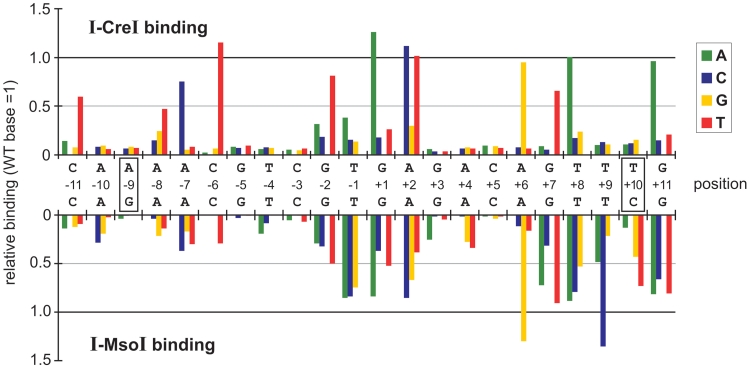
Figure 2.
In vitro binding specificity profiles of I-CreI and I-MsoI. The relative binding affinities of I-CreI (top) and I-MsoI (bottom) for all 4 bp at each target site position were determined using a competitive in vitro binding assay (20). Target site base pair numbering is as in Figure 1, with positions differing between I-CreI and I-MsoI shown in boxes (−9 and +10 positions). Bar heights indicate the binding affinity of each protein for base pair substitutions relative to binding of the native base pair whose binding has been arbitrarily set as 1.0. All results are the mean of three replications in which the standard deviation between experiments was ±5%. Error bars have been omitted for graphical clarity.
The 10 bp positions in the I-CreI target site (±3–5 and ±9–10) that were palindromically symmetric between the left and right half sites displayed a strong preference to bind native target site base pairs, and greatly reduced affinities (≤10% of native) for each of the other 3 bp (Figure 2, top panel). Base pairs at these positions make multiple direct or water-mediated contacts (17,18). In contrast, the central four target site positions (−2 to +2) that make no base-specific contacts with I-CreI were able to bind 1 or 2 bp in addition to the native base pair with affinities of ≥50% that of the native base pair. Seven of the eight remaining I-CreI target site positions (±6–7, +8 and ±11) each bound at least one variant base pair with affinities of ≥50% that of the native base pair (Figure 2, top panel). Only 7 of 66 single base pair substitutions in the I-CreI target site displayed binding affinities comparable to the native base pair (−6C > T, +1G > A, +2A > C or T, +6A > G, +8T > A and +11G > A, Figure 2, top panel). Of note, many base pair variants with high binding affinities increased the overall symmetry of the I-CreI target site (see, e.g. −7A > C and +7G > T; −6C > T and +6A > G, Figure 2, top panel).
The in vitro binding profile of I-MsoI differed substantially from I-CreI and was less specific. I-MsoI had only 4 bp positions (versus I-CreI's 10), positions −9, ±5 and −3, with a strong preference for binding only the native base pair (Figure 2, bottom panel). I-MsoI was also more tolerant of base pair changes in the central four target site positions (−2 to +2), where all three alternative base pairs could be bound with affinities ranging from 30 to 90% of the native base pair (Figure 2, bottom panel). Again, base pair substitutions that increased overall target site symmetry, e.g. at positions +6 and +9, increased site binding affinity (Figure 2, bottom panel).
A conspicuous difference between I-MsoI and I-CreI was site binding symmetry: the left I-MsoI half site (−3 to −11) displayed higher binding specificity than did I-CreI, whereas the right I-MsoI half site had six positions (+6 to +11) where one or more variant base pair was bound with ≥50% of the affinity of the native base pair. This difference in I-MsoI half site binding affinities agrees with our prior analysis of the I-MsoI DNA binding thermodynamic profile (24).
We used these site binding data to calculate global binding specificities for I-CreI and I-MsoI. The binding specificity of I-CreI was ∼1.8 × 10−10, which was calculated by dividing the number of variant target sites that were bound with ≥50% of native site affinity (n = 3072) by the number of unique target sites of length 22 bp (n = 422 = 1.8 × 1013). The corresponding binding specificity of I-MsoI was approximately an order of magnitude lower, or ∼1.6 × 10−9. The corresponding binding specificity profiles of mCreI and mMsoI closely resembled their respective parental proteins (Supplementary Figure S1), although their calculated binding specificities were lower: ∼1.6 × 10−9 for mCreI, and ∼2.0 × 10−6 for mMsoI. This difference may reflect the presence of the 33 residue linkers inserted to monomerize I-CreI and I-MsoI (Figure 1) (19), and/or the presence of two His tags on the subunits of the homodimeric proteins as opposed to the single tag on each corresponding monomeric protein in binding assays.
In vitro cleavage specificity
In vitro cleavage specificities were determined using a single tube, competitive ‘bar code’ cleavage assay to simultaneously assess the cleavage sensitivity of all 4 bp possibilities at each target site position (Figure 3A) (25). Target site libraries were constructed in pDR-GFPuniv (http://depts.washington.edu/monnatws/plasmids/pDR-GFP%20univ.pdf) to permit the same target site libraries to be used for in vitro and in vivo cleavage specificity determinations (Figure 4; see below). In vitro cleavage conditions were determined to ensure ∼50% cleavage of the native target site at a 1:1 protein:DNA ratio. This ratio was chosen to minimize the effect of binding affinity differences upon cleavage. The results of 22 competitive cleavage reactions that simultaneously assayed all 4 bp possibilities at each base pair position were then displayed on a single gel for quantitation (Figure 3B).
I-CreI displayed a strong preference for native base pairs at many target site positions (±1 to ±10) in cleavage assays (Figure 3B and C, top panels). The highest cleavage specificities were observed at target site positions ±3–4 and ±9–10, and the lowest specificities at positions ±1, 2 and 8. Only 5 bp substitutions were cleaved more efficiently than the native base pair: −7A > C, −2G > A, +1G > A, +7G > T and +8T > A. Two of these substitutions, −7A > C and +7G > T, increased the overall symmetry of the I-CreI/mCreI target site.
I-MsoI displayed the highest cleavage specificity at target site positions ±3 and ±5, and was least specific at target site positions −1 and +7, 8 and 11 (Figure 3B and C, bottom panels). At positions −1 and +11, I-MsoI cleaved all 4 bp possibilities with equal efficiency. A total of 14 bp substitutions at seven target site positions were cleaved, as well as the corresponding native base pair: −7A > T, −1T > A/C/G, +2A > C, +7G > A/C/T, +8T > A/C, +10C > T and +11G > A/C/T. Four of these substitutions, −1T > C, +2A > C, +7G > T and +10C > T, increased the overall symmetry of the I-MsoI target site (Figure 3C, bottom panel).
The global cleavage specificities of I-CreI and I-MsoI were calculated from the number of variant target sites that could be cleaved with ≥50% of the efficiency of the native site, divided by the number of unique 20 bp (I-CreI) or 22 bp (I-MsoI) target sites. These cleavage specificities were ∼1.4 × 10−8 for I-CreI and ∼5.4 × 10−5 for I-MsoI. The corresponding in vitro cleavage specificity profiles for mCreI and mMsoI were very similar to I-CreI and I-MsoI: respectively ∼2.8 × 10−8 and ∼2.4 × 10−5 (Supplementary Figure S2). The higher global binding and cleavage specificities of I-CreI versus I-MsoI can be seen easily in relative binding and cleavage difference plots that compare the two endonucleases (Supplementary Figure S3).
In vitro cleavage of target sites with multiple base pair changes
In order to determine whether single base pair cleavage data could be used to predict the cleavage sensitivity of target sites having multiple base pair changes, we analyzed 36 different mCreI target sites containing from 3 to 9 bp differences from the native I-CreI target site (Figure 1). An explicit example from these analyses is shown in Figure 5, in which our mCreI cleavage specificity profiles were used to search for engineerable target sites in the human SBDS gene to target to catalyze gene repair.
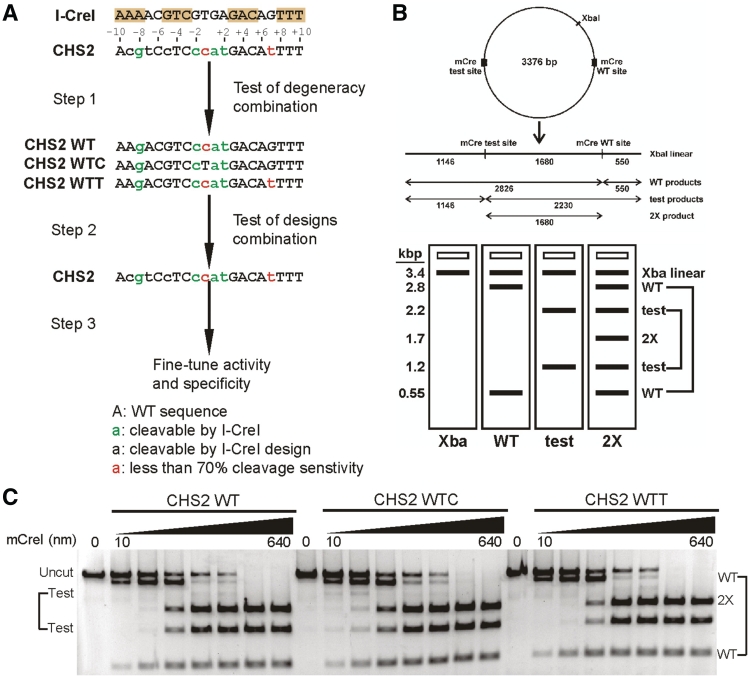
Figure 5.
In vitro cleavage specificity profiling guides the generation of target site-specific LHE variants. (A) The workflow for engineering mCreI towards novel target sites, using the human Shwachman-Diamond syndrome gene SBDS CHS2 site as an example. The first step is to predict the cleavage sensitivity of target sites containing multiple base pair changes from single base pair cleavage sensitivity profiling data (see, e.g. Figure 2). These predictions can be experimentally verified in a second step, and then combined with LHE protein designs to generate a target site-specific LHE variant. The activity and specificity of this novel target site-specific variant can then be further improved by a combination of selection or screening. (B) Schematic overview of in vitro competitive cleavage assay. Both native and novel target sites are cloned into a plasmid that is linearized prior to LHE cleavage. Cleaved products are visualized on an agarose gel to determine the relative cleavage efficiency of the native and test sites from relative band intensities. (C) Agarose gels displaying in vitro cleavage efficiency of mCreI on three CHS2 sites that contain different combinations of base pair changes.
SBDS-inactivating mutations cause Shwachman-Diamond syndrome (SDS), a rare, heritable bone marrow failure syndrome characterized by congenital abnormalities, hematopoietic failure and cancer predisposition (26). The human SBDS CHS2 mCreI target site is located in SBDS intron 1, upstream of the location of >90% of SDS-causing SBDS mutations (27). The CHS2 SBDS site differs at 9 bp positions from the native I-CreI target site (Figure 5A). Our in vitro cleavage data and a prior systematic protein computation design analysis of mCreI (25) indicated that four of these base pair differences should be recognized and cleaved by native I-CreI/mCreI (−8, −1, +1 and +2), and an additional 3 bp changes at positions −9, −7 and −5 could be successfully targeted by previously identified mCreI protein computational designs (25). The remaining 2 bp differences, at positions −1 and +7, were predicted to reduce cleavage.
Cleavage analysis of CHS2 site variants containing different combinations of these base pair changes allowed us to verify the predictions of cleavage sensitivity for the combined base pair differences at positions −8, −1, +1 and +2, and that substitutions at −1 and +7 reduced though did not abolish cleavage. Similar analyses of 35 additional target sites chosen on the basis of cleavage degeneracy data and engineerability with from 3 to 7 bp differences from the native I-CreI target site revealed that a majority (31/35, or 89%) predicted to be cleavage-sensitive from our single base pair scanning data were cleavage-sensitive, and that 11 of these sites (31%) were cleaved with efficiencies comparable to the native I-CreI target site. Many target sites with up to three contiguous base pair changes, each having relative cleavage activities of >0.5 versus the native site were cleavage-sensitive as predicted, whereas target sites having four or five contiguous substitutions where one or two substitutions had relative cleavage activities of ≤0.5 in our single base pair scan data were largely cleavage-resistant (additional results not shown).
Using single base pair scan data to predict the potential for evolutionary spread
We also used our single base pair cleavage data to gauge the potential of I-CreI or I-MsoI for lateral transfer to additional organisms to identify target site variants that retained 28S rRNA secondary structure motifs required for function (the extrahelical +6A base and a paired stem-loop structure; Figure 6A), and were predicted to be highly cleavage sensitive from our target site scan results. The I-CreI site predicted to be the most cleavage-sensitive by these criteria had −7C and +8G base pair substitutions. Blastn searches using this site identified 75 perfect matches in nucleotide sequence databases, of which 55 were in LSU ribosomal RNA genes. Two I-MsoI sites with predicted higher cleavage sensitivity, in contrast, did not have perfect matches that could be identified by Blast searching (Supplementary Table S1; additional results not shown).
Figure 6.
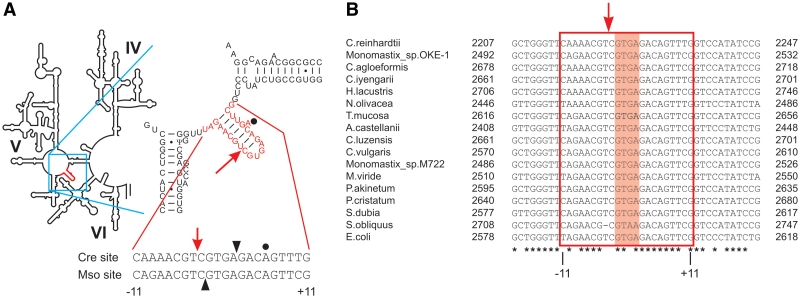
Location of I-CreI and I-MsoI target sites in host ribosomal RNA genes. (A) The secondary structure of domain V in E. coli 23S rRNA gene, in which the n.2593 target site region recognized by I-CreI and I-MsoI is shown in red and the intron insertion site indicated by an arrow. The I-CreI and I-MsoI target sites are aligned below the stem-loop to indicate intron insertion sites (down arrow), strand cleavage sites (filled arrow heads) and the location of the extrahelical base at the +6 position that plays a role in peptide release (dot) (38). (B) Aligned sequences of the corresponding n.2593 target site region from LSU rRNA genes from chloroplast or mitochondrial 23S ribosomal RNA genes of green algae, with the E. coli 23S ribosomal RNA gene shown at bottom (33). Target sites are surrounded by a red box, with the central four positions (−2 to +2) shaded in magenta. Positions conserved across all target sites are underlined with an asterisk.
In vivo cleavage specificity
In light of growing interest in using HE proteins for genome engineering and gene therapy, we also determined the cleavage specificities of mCreI and mMsoI in human cells. These experiments used the same target site libraries constructed for in vitro cleavage specificity experiments (Figure 3). Target site cleavage of site plasmids in vivo was quantified from the frequency of cleavage-dependent generation of recombinant, GFP+ cells (Figure 4A) (19). In vivo cleavage specificity profiles for mCreI and mMsoI closely resembled those determined in vitro (Figures 3C and 4B). One conspicuous difference was the overall higher specificity of mMsoI cleavage in vivo at right half site positions +7 to +11. In contrast, mCreI in vitro and in vivo cleavage specificity profiles closely resembled one another (Figures 3C and 4B).
DISCUSSION
Relationship of in vitro binding and cleavage specificities
We observed a strong correlation between binding and cleavage at many I-CreI and I-MsoI target site positions. Most substitutions that reduced binding also comparably reduced cleavage efficiency (Figures 2 and 3). Of greater interest were base pair substitutions that disproportionately affected binding or cleavage: these substitutions may provide insight into dynamic aspects of binding and cleavage complex formation. Four I-CreI site substitutions substantially reduced binding, but retained >50% of the cleavage activity of the native target site (−8A > C or G, −5G > T and −2G > A). One substitution, +2A > T, displayed native binding affinity but no detectable cleavage activity. I-MsoI had 15 bp substitutions that disproportionately reduced binding versus cleavage. One I-MsoI substitution, +9T > C, reduced cleavage by ∼40%, while enhancing binding to >100% (Figures 2 and 3). These ‘uncoupling’ base pair substitutions are easy to identify in difference plots that compare binding and cleavage activity at each target site base pair position (Supplementary Figure S4).
Base pair substitutions that selectively affect binding but not cleavage may allow transition state complexes to be formed that include new stabilizing interactions, or that do not depend on stabilizing interactions in the ground state. Conversely, base pair substitutions that selectively affect cleavage but not binding might create interactions that stabilized the ground state, or that hindered conformational changes required to form a transition state complex. It may be possible to discriminate among these models by determining the predicted binding energies or structures of specific ‘uncoupling’ substitutions captured in both pre-cleavage and cleavage complexes.
An important determinant of the higher overall specificity of I-CreI is the larger number of direct and water-mediated contacts made with target site DNA: I-CreI makes an average of 2.4 contacts (direct or water-mediated) with each target site position, whereas I-MsoI makes an average of only 1.7 contacts/bp (18). The most specific target site positions in both I-CreI and I-MsoI, e.g. the ±3–5 target site positions (Figures 2 and 3), share many conserved DNA-protein contacts that may be required in both proteins to correctly position target site DNA to promote cleavage (Figure 1C) (16,18). Additional DNA-protein contacts at positions more distant from the active sites, e.g. positions ±6–11, help ensure high site binding affinity and sequence-specific discrimination.
Target site symmetry also plays an important role in determining overall site specificity and, as we discuss below, is likely to be an important constraint on both lateral transfer and HE evolution. The importance of site symmetry is most clearly revealed by base pair substitutions that create a higher degree of I-CreI or I-MsoI target site symmetry: these substitutions almost invariable lead to enhanced target site binding and/or cleavage [see, e.g. (24)].
Site specificity profiling enables HE-mediated genome engineering applications
HE site specificity profiles provide useful information to guide the generation of HE variants for genome engineering (6,28–31). Position-specific search or scoring matrices (PSSMs) can be constructed from profiling data such as those presented in Figure 2, and used to identify gene-specific or genomic target sites that have a high likelihood of being bound or cleaved by specific HEs. The functional consequences of base pair differences including SNP variants in ‘near match’ sites can be predicted from profiling data, as shown in Figure 5. The most important target site positions on which to focus specificity engineering efforts can be identified early, as can genomic target sites where there are few or no DNA contacts to modify to achieve higher or altered specificity (e.g. in the central four target site positions −2 to +2). Target sites likely to be confounded by low specificity or the potential for substantial off-target cleavage activity can also be identified and avoided where there are better alternatives. These results provide a good example of how single base pair profiling data can be used to determine engineering feasibility, and focus protein engineering on specific base pair positions where protein engineering is required to achieve new site specifity.
I-CreI and I-MsoI site specificity versus host target gene structure
Many HE ORFs are found as open reading frames in large or small subunit ribosomal RNA genes (the LSU/23/25/28S and SSU/16S/18S rRNA genes) (16,32). The native I-CreI/I-MsoI LSU target site resides in a highly conserved segment of the chloroplast LSU genes of Chlamydomonas and Monomastix (LSU n.2593, where nucleotide numbering is keyed to the reference Escherichia coli LSU 23S ribosomal RNA gene sequence) (33). The corresponding portion of LSU rRNA is located in the central loop of domain V that includes the peptidyl transferase center (Figure 6A) (34–36). Nucleotides flanking the LSU n.2593 insertion site display 2-fold symmetry, and form a stem-loop structure in rRNA secondary structure models. The stem in structural models is formed by base pairs at nucleotide positions n.2588–2594 (corresponding to Cre/Mso target site positions −9 to −3) and n.2599–2606 [corresponding to target site positions +3 to +10; Figures 1 and 6; (37)]. The four nucleotides between the two-half sites, n.2595–2599, form an unpaired loop in rRNA that corresponds to the central four nucleotides in the Cre/Mso target site (positions −2 to +2, Figures 1 and 6A). The A residue at position n.2602, corresponding to position +6 within the Cre/Mso target site, is extrahelical in RNA secondary structure models and has been shown to be essential for ribosomal peptide release (38).
The n.2588–2606 stem-loop region of LSU rDNA thus provides a well-defined and highly conserved target for the lateral transfer of HE-encoding mobile introns. The binding and cleavage specificity profiles of I-CreI and I-MsoI reflect and exploit these LSU target site constraints. LSU bases that form the stem-loop structure in LSU rRNA correspond to Cre/Mso target site positions −9 to −3, +3 to +5 and +7 to +10 (Figure 6A), where I-CreI and I-MsoI display high binding and cleavage specificity (Figures 2 and 3). The central 4 bp positions (−2 to +2) in both target sites, in contrast, are located in a loop with few or no apparent sequence constraints in rRNA secondary structure models, and these positions contribute little target site binding or cleavage specificity (Figures 2 and 3).
Implications for HE evolution
The near-perfect 2-fold symmetry of the n.2593 LSU target site is dictated by rRNA functional constraints. These constraints, in turn, may strongly influence HE protein evolution at several levels. One potential advantage of using a highly conserved, largely symmetric target site for homing is that symmetric sites can be effectively targeted by small, homodimeric HE proteins encoded by a single, short open reading frame. This permits homing to be mediated by the lateral transfer of a small open reading frame and accompanying intron or intein that is easily and reliably transferred. Small mobile intron/intein open reading frames also present a small target for potentially inactivating mutations.
Duplication or duplication and fusion of an open reading frame encoding a homodimeric LHE subunit opens up another evolutionary opportunity: the ability to target asymmetric, degenerate or non-palindromic target sites. This strategy can be glimpsed in the structure of I-MsoI, a symmetric homodimeric LHE which uses asymmetric contacts to recognize a target site with a high degree of target site symmetry (18). Another particularly instructive example is I-CeuI, an asymmetric, homodimeric LHE from Chlamydomonas eugametos that uses unique structural elaborations on the core LHE fold to cleave the highly asymmetric n.1923 LSU target site in C. eugamotes chloroplast DNA. Of note, I-CeuI retains cleavage activity on symmetric-left or symmetric-right target sites (39). The ability to cleave related symmetric and asymmetric target sites could broaden the range of potential LHE hosts to include organisms with related asymmetric, as well as symmetric, target sites.
The substantially different structural solutions used by I-CreI, I-MsoI and I-CeuI to target LSU sites with differing degrees of asymmetry suggest two different evolutionary strategies that may ensure the evolutionary persistence of HE proteins and their encoding selfish DNA elements. I-CreI, with a rich set of DNA-protein contacts, can discriminate between closely related target sites (17,18,39). A potential advantage of this higher site specificity is the ability to evolve higher cleavage activity to aid lateral transfer, without substantially increasing cleavage-dependent host toxicity (19). I-MsoI and I-CeuI, in contrast, may be able to spread to a wider range of new hosts by virtue of less stringent target site sequence requirements. The potentially deleterious consequences of lower site specificity may be offset by lower cleavage activity, as is the case for I-MsoI. Either of these strategies for coupling site recognition specificity and cleavage activity could represent a viable—or preferred—strategy for lateral transfer and evolutionary persistence in host populations with differing degrees of target site sequence divergence.
Host accommodation following lateral transfer represents another important determinant of HE evolution. Several strategies for host accommodation have been identified among HEs. These include use of an HE-encoded maturase function to improve the expression of host genes; host genetic code adoption and the use of host codon preferences to improve HE expression; and modulation of HE protein expression to ensure adequate expression of both host gene and HE open reading frame-encoded proteins (6,40,41). It should be possible to experimentally determine the contribution of these determinants of HE lateral transfer using the systems that have been developed to study homing and LAGLIDADG HEs in prokaryotes (22,42,43), single cell eukaryotes (44,45), and most recently metazoans (46,47).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures S1–S4 and Supplementary Table S1.
FUNDING
U.S. National Institutes of Health Training Grant (5RL9HL092555 to H.L.); U.S. National Institutes of Health Interdisciplinary Training Grant in Cancer Research (T32 CA80416 to U.Y.U.); U.S. National Institutes of Health F30 Fellowship Award (DK083223 to U.Y.U.); Poncin Fund Awards (to U.Y.U.); U.S. National Institutes of Health U54 Interdisciplinary Research Roadmap Award (1RL1 CA133831 to R.J.M. Jr); and Bill and Melinda Gates Foundation/Foundation for the National Institutes of Health Grand Challenges in Global Health Awards (to R.J.M. Jr). Funding for open access charge: National Institutes of Health U54 Interdisciplinary Research Roadmap Award (1RL1 CA133831 to R.J.M. Jr).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Alden Hackman for assistance with figure preparation, Barry Stoddard and his lab for many useful discussions and manuscript suggestions, Akiko Shimamura and her lab for help in developing the human SBDS gene targeting analysis and Shelia Teves for help with in vivo cleavage assays.
REFERENCES
- 1.Dujon B. Group I introns as mobile genetic elements: facts and mechanistic speculations – a review. Gene. 1989;82:91–114. doi: 10.1016/0378-1119(89)90034-6. [DOI] [PubMed] [Google Scholar]
- 2.Lambowitz AM, Belfort M. Introns as mobile genetic elements. Annu. Rev. Biochem. 1993;62:587–622. doi: 10.1146/annurev.bi.62.070193.003103. [DOI] [PubMed] [Google Scholar]
- 3.Belfort M, Perlman PS. Mechanisms of intron mobility. J. Biol. Chem. 1995;270:30237–30240. doi: 10.1074/jbc.270.51.30237. [DOI] [PubMed] [Google Scholar]
- 4.Belfort M. Back to basics: structure, function, evolution and application of homing endonucleases and inteins. In: Belfort M, Stoddard BL, Wood DW, Derbyshire V, editors. Homing Endonucleases and Inteins. Berlin: Springer-Verlag; 2005. pp. 1–10. [Google Scholar]
- 5.Stoddard BL. Homing endonuclease structure and function. Q. Rev. Biophys. 2005;38:1–47. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 6.Stoddard BL. Homing endonucleases: from microbial genetic invaders to reagents for targeted DNA modification. Structure. 2011;19:7–15. doi: 10.1016/j.str.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chevalier B, Monnat RJ, Jr, Stoddard BL. The LAGLIDADG homing endonuclease family. In: Belfort M, Stoddard BL, Wood DW, Derbyshire V, editors. Homing Endonucleases and Inteins. Springer-Verlag, Berlin: 2005. pp. 33–47. [Google Scholar]
- 8.Grishin A, Fonfara I, Alexeevski A, Spirin S, Zanegina O, Karyagina A, Alexeyevsky D, Wende W. Identification of conserved features of LAGLIDADG homing endonucleases. J. Bioinform. Computat. Biol. 2010;8:453–469. doi: 10.1142/s0219720010004665. [DOI] [PubMed] [Google Scholar]
- 9.Colleaux L, D'Auriol L, Galibert F, Dujon B. Recognition and cleavage site of the intron-encoded omega trnasposase. Proc. Natl Acad. Sci. USA. 1988;85:6022–6026. doi: 10.1073/pnas.85.16.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Argast GM, Stephens KM, Emond MJ, Monnat RJ., Jr I-PpoI and I-CreI homing site sequence degeneracy determined by random mutagenesis and sequential in vitro enrichment. J. Mol. Biol. 1998;280:345–353. doi: 10.1006/jmbi.1998.1886. [DOI] [PubMed] [Google Scholar]
- 11.Gimble FS. Invasion of a multitude of genetic niches by mobile endonuclease genes. FEMS Microbiol. Lett. 2000;185:99–107. doi: 10.1111/j.1574-6968.2000.tb09046.x. [DOI] [PubMed] [Google Scholar]
- 12.Burt A, Koufopanou V. Homing endonuclease genes: the rise and fall and rise again of a selfish element. Curr. Opin. Genet. Dev. 2004;14:609–615. doi: 10.1016/j.gde.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 13.Burt A, Trivers R. Genes in Conflict: The Biology of Selfish Genetic Elements. Cambridge, MA: The Belknap Press of Harvard University Press; 2006. [Google Scholar]
- 14.Goddard MR, Burt A. Recurrent invasion and extinction of a selfish gene. Proc. Natl Acad. Sci. USA. 1999;96:13880–13885. doi: 10.1073/pnas.96.24.13880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thompson AJ, Yuan X, Kudlicki W, Herrin DL. Cleavage and recognition pattern of a double-strand-specific endonuclease (I-CreI) encoded by the chloroplast 23S rRNA intron of Chlamydomonas reinhardtii. Gene. 1992;119:247–251. doi: 10.1016/0378-1119(92)90278-w. [DOI] [PubMed] [Google Scholar]
- 16.Lucas P, Otis C, Mercier JP, Turmel M, Lemieux C. Rapid evolution of the DNA-binding site in LAGLIDADG homing endonucleases. Nucleic Acids Res. 2001;29:960–969. doi: 10.1093/nar/29.4.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jurica MS, Monnat RJ, Jr, Stoddard BL. DNA recognition and cleavage by the LADLIDADG homing endonuclease I-CreI. Mol. Cell. 1998;2:469–476. doi: 10.1016/s1097-2765(00)80146-x. [DOI] [PubMed] [Google Scholar]
- 18.Chevalier B, Turmel M, Lemieux C, Monnat RJ, Stoddard BL. Flexible DNA target site recognition by divergent homing endonuclease isoschizomers I-CreI and I-MsoI. J. Mol. Biol. 2003;329:253–269. doi: 10.1016/s0022-2836(03)00447-9. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Pellenz S, Ulge U, Stoddard BL, Monnat RJ., Jr Generation of single-chain LAGLIDADG homing endonucleases from native homodimeric precursor proteins. Nucleic Acids Res. 2009;37:1650–1662. doi: 10.1093/nar/gkp004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao L, Pellenz S, Stoddard BL. Activity and specificity of the bacterial PD-(D/E)XK homing endonuclease I-Ssp6803I. J. Mol. Biol. 2009;385:1498–1510. doi: 10.1016/j.jmb.2008.10.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doyon JB, Pattanayak V, Meyer CB, Liu DR. Directed evolution and substrate specificity profile of homing endonuclease I-SceI. J. Am. Chem. Soc. 2006;128:2477–2484. doi: 10.1021/ja057519l. [DOI] [PubMed] [Google Scholar]
- 23.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eastberg JH, Smith AM, Zhao L, Ashworth J, Shen BW, Stoddard BL. Thermodynamics of DNA target site recognition by homing endonucleases. Nucleic Acids Res. 2007;35:7209–7221. doi: 10.1093/nar/gkm867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ulge UY, Baker D, Monnat RJ., Jr Comprehensive computational design of mCreI homing endonuclease cleavage specificity for genome engineering. Nucleic Acids Res. 2011;39:4330–4339. doi: 10.1093/nar/gkr022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burroughs L, Woolfrey A, Shimamura A. Shwachman-Diamond syndrome: a review of the clinical presentation, molecular pathogenesis, diagnosis, and treatment. Hematol. Oncol. Clin. North Am. 2009;23:233–248. doi: 10.1016/j.hoc.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boocock GRB, Morrison JA, Popovic M, Richards N, Ellis L, Durie PR, Rommens JM. Mutations in SBDS are associated with Shwachman-Diamond syndrome. Nat. Genet. 2003;33:97–101. doi: 10.1038/ng1062. [DOI] [PubMed] [Google Scholar]
- 28.Belfort M, Roberts RJ. Homing endonucleases: keeping the house in order. Nucleic Acids Res. 1997;25:3379–3388. doi: 10.1093/nar/25.17.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arnould S, Chames P, Perez C, Lacroix E, Duclert A, Epinat JC, Stricher F, Petit AS, Patin A, Guillier S, et al. Engineering of large numbers of highly specific homing endonucleases that induce recombination on novel DNA targets. J. Mol. Biol. 2006;355:443–458. doi: 10.1016/j.jmb.2005.10.065. [DOI] [PubMed] [Google Scholar]
- 30.Smith J, Grizot S, Arnould S, Duclert A, Epinat JC, Chames P, Prieto J, Redondo P, Blanco FJ, Bravo J, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Redondo P, Prieto J, Munoz IG, Alibes A, Stricher F, Serrano L, Cabaniols JP, Daboussi F, Arnould S, Perez C, et al. Molecular basis of xeroderma pigmentosum group C DNA recognition by engineered meganucleases. Nature. 2008;456:107–111. doi: 10.1038/nature07343. [DOI] [PubMed] [Google Scholar]
- 32.Haugen P, Simon DM, Bhattacharya D. The natural history of group I introns. Trends Genet. 2005;21:111–119. doi: 10.1016/j.tig.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 33.Brosius J, Dull TJ, Noller HF. Complete nucleotide sequence of the 23S ribosomal RNA gene from Escherichia coli. Proc. Natl Acad. Sci. USA. 1980;77:201–204. doi: 10.1073/pnas.77.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dunkle JA, Cate JH. Ribosome structure and dynamics during translocation and termination. Annu. Rev. Biophys. Biomol. Struct. 2010;39:227–244. doi: 10.1146/annurev.biophys.37.032807.125954. [DOI] [PubMed] [Google Scholar]
- 35.Ramakrishnan V. Unraveling the structure of the ribosome (Nobel Lecture) Angewandte Chemie International Edition. 2010;49:4355–4380. doi: 10.1002/anie.201001436. [DOI] [PubMed] [Google Scholar]
- 36.Moore PB, Steitz TA. The roles of RNA in the synthesis of protein. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a003780. pii:a003780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galburt EA, Chevalier B, Tang W, Jurica MS, Flick KE, Monnat RJ, Jr, Stoddard BL. A novel endonuclease mechanism directly visualized for I-PpoI. Nat. Struct. Biol. 1999;6:1096–1099. doi: 10.1038/70027. [DOI] [PubMed] [Google Scholar]
- 38.Sato NS, Hirabayashi N, Agmon I, Yonath A, Suzuki T. Comprehensive genetic selection revealed essential bases in the peptidyl-transferase center. Proc. Natl Acad. Sci. USA. 2006;103:15386–15391. doi: 10.1073/pnas.0605970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spiegel PC, Chevalier B, Sussman D, Turmel M, Lemieux C, Stoddard BL. The structure of I-CeuI homing endonuclease: evolving asymmetric DNA recognition from a symmetric protein scaffold. Structure. 2006;14:869–880. doi: 10.1016/j.str.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 40.Gogarten JP, Hilario E. Inteins, introns, and homing endonucleases: recent revelations about the life cycle of parasitic genetic elements. BMC Evol. Biol. 2006;6:94. doi: 10.1186/1471-2148-6-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stoddard B, Belfort M. Social networking between mobile introns and their host genes. Mol. Microbiol. 2010;78:1–4. doi: 10.1111/j.1365-2958.2010.07217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seligman LM, Stephens KM, Savage JH, Monnat RJ., Jr Genetic analysis of the Chlamydomonas reinhardtii I-CreI mobile intron homing system in Escherichia coli. Genetics. 1997;147:1653–1664. doi: 10.1093/genetics/147.4.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gimble FS, Moure CM, Posey KL. Assessing the plasticity of DNA target site recognition of the PI-SceI homing endonuclease using a bacterial two-hybrid selection system. J. Mol. Biol. 2003;334:993–1008. doi: 10.1016/j.jmb.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 44.Jacquier A, Dujon B. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell. 1985;41:383–394. doi: 10.1016/s0092-8674(85)80011-8. [DOI] [PubMed] [Google Scholar]
- 45.Gimble FS, Thorner J. Homing of a DNA endonuclease gene by meiotic gene conversion in Saccharomyces cerevisiae. Nature. 1992;357:301–306. doi: 10.1038/357301a0. [DOI] [PubMed] [Google Scholar]
- 46.Windbichler N, Papathanos PA, Catteruccia F, Ranson H, Burt A, Crisanti A. Homing endonuclease mediated gene targeting in Anopheles gambiae cells and embryos. Nucl. Acids Res. 2007;35:5922–5933. doi: 10.1093/nar/gkm632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Windbichler N, Menichelli M, Papathanos PA, Thyme SB, Li H, Ulge UY, Hovde BT, Baker D, Monnat RJ, Burt A, et al. A synthetic homing endonuclease-based gene drive system in the human malaria mosquito. Nature. 2011;473:212–215. doi: 10.1038/nature09937. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.