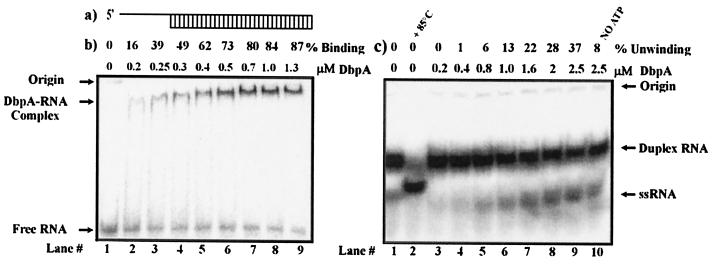
Figure 1.
Binding and unwinding activity of DbpA with short dsRNA. (a) Schematic of the short dsRNA substrate used for the following assays. (b) Gel retardation assay of DbpA and dsRNA substrate. Binding reactions were performed in a 20-μl reaction volume in the presence of 5 pmol of 32P-labeled dsRNA and increasing amounts of DbpA. The binding percentages are indicated. Free RNA, RNA-DbpA complexes, and the origins of the wells in the gel are indicated on the left. (c) Unwinding of dsRNA by DbpA. Lane 1 represents free dsRNA. Lane 2 shows the ssRNA that resulted after 3 min of incubation at 85°C. Lanes 3–10 show the unwinding reaction at a given protein concentration. The differences in the position of ssRNA on the gel between lane 2 and lanes 3–10 are caused by differences in ionic strength of the free dsRNA used for control and the product of the enzymatic reaction.