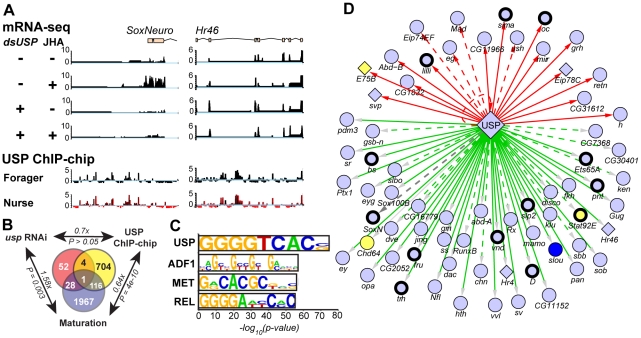
Figure 2. Putative direct and indirect targets of USP in honey bee fat bodies.
Putative targets of USP in the fat bodies were characterized both by usp RNAi—deep mRNA sequencing (in combination with juvenile hormone analog, JHA, treatments) as well as by USP ChIP-chip with fat body tissue samples from nurses and from foragers. A. Genomic regions surrounding two putative target genes, the transcription factors SoxNeuro and Hr46. Units for mRNA-seq are read counts, and for ChIP-chip the ratio of a-USP to control. B. Venn diagram shows that many USP target genes identified by usp RNAi and USP ChIP-chip are differentially expressed between nurses and foragers (“Maturation”). Fold enrichment of overlap and its significance (hypergeometric test) are indicated for each comparison. C. The 1360 genomic binding sites of USP are enriched for conserved cis-regulatory sequences, putatively recognized by the TFs shown at left. D. USP binds genomic locations near 67 transcription factors (TFs), including members of the nuclear hormone receptor family (diamonds) and other TF families (circles). Some of these TFs were differentially expressed between nurses and foragers (blue, higher in nurse; yellow, higher in forager), and predicted targets for several of these TFs based on transcriptional regulatory network analysis [20] were enriched for genes that are differentially expressed in maturation-related contexts (thick outlines). Some of these TFs were also identified as USP targets in D. melanogaster [55] (red lines), and some binding sites contain the GGGGTCACS cis-regulatory sequence recognized by USP in D. melanogaster (solid lines).