Figure 5. cis-regulatory sequences predict behaviorally-related responses of USP targets.
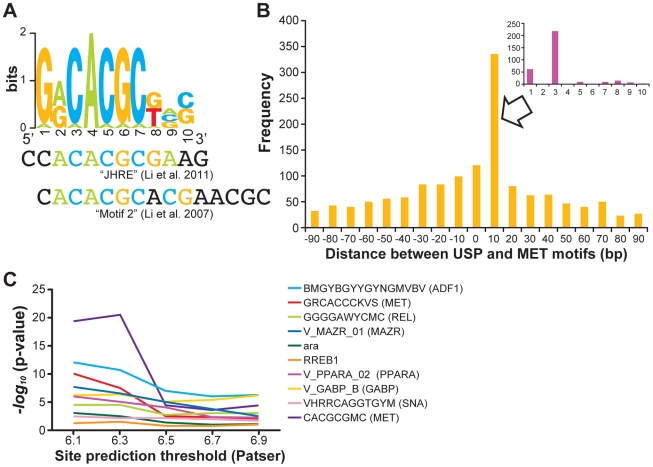
A. The GRCACGCKVS motif enriched at USP binding sites (Figure 2C) matches a Juvenile Hormone Response Element (JHRE) recognized by MET (and other bHLH TFs) [24], [61]. B. Predicted binding sites of USP and MET (the GGGGTCACS and GRCACGCKVS motifs, respectively) were consistently overlapping, with start positions located 3 bp apart. All adjacent pairs of Patser-predicted matches to these motifs in USP-binding loci (ChIP peaks) were considered; shown is the histogram of spacing between start positions of each pair, with negative numbers indicating that the MET site is on the left. Inset shows a zoomed-in view of the histogram for spacings in the range of 1–10 bp. C. Spacing constraints between sites of GGGGTCACS (USP) and potential cofactor motifs. Each of the 10 most significantly overrepresented motifs in USP-binding loci was tested for a constraint on spacing (< = 25 bp vs. >25 bp) between sites for USP and that motif. The Y-axis shows the −log10 of the p-value of this test, at five different statistical thresholds (X-axis) for defining motif matches.