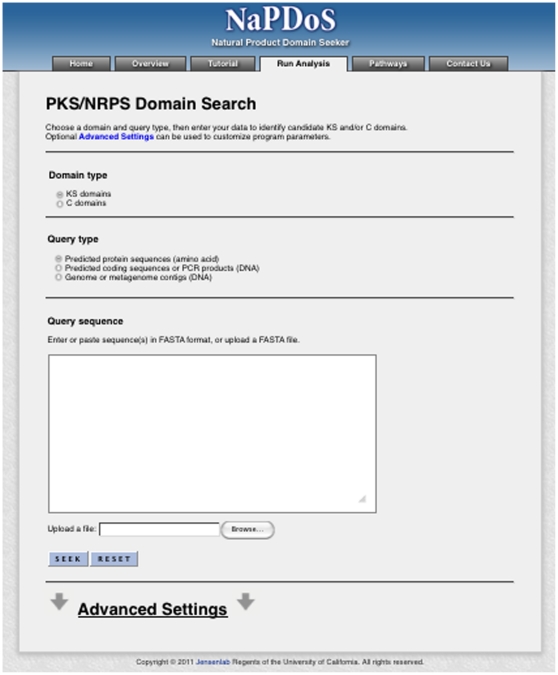
Figure 1. NaPDoS bioinformatic pipeline.
The web interface to this pipeline is divided 3 consecutive steps. Nucleic acid sequences are translated into predicted amino acids and genomic sequences are screened using Hidden Markov Models (HMM). For protein and small nucleic acid sequences a BLAST search is performed against curated reference database examples to identify matches to known PKS/NRPS pathways. Selected candidate sequences plus the BLAST results are trimmed and inserted into a manually curated reference alignment, keeping the original reference alignment intact. This alignment is used to build a tree.