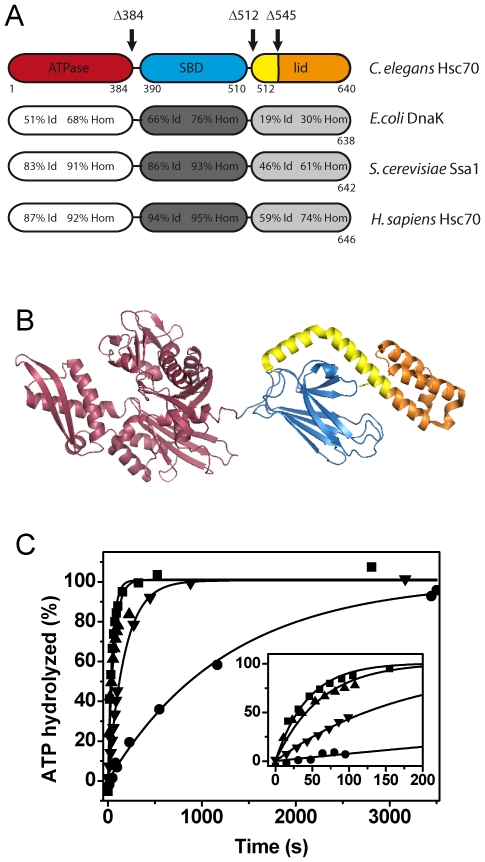
Figure 2. CeHsc70 truncation mutants show an altered ATP turnover.
(A) Domain organization and amino acid identity (Id) and homology (Hom) of CeHsc70 towards bacterial, yeast and human homologs. The truncation mutants generated in this work are indicated by black arrows. (B) Structure of DnaK based on the PDB file 2KHO [93]. The truncations are colored in red (CeHsc70-Δ384), red and blue (CeHsc70-Δ512) and red, blue and yellow (CeHsc70-Δ545). The lid region, which is missing in the CeHsc70-Δ545 mutant, is highlighted in orange. (C) The single-turnover experiments using 20 µM CeHsc70 variants were performed as outlined in the Materials and Methods section in standard buffer at 25°C. Data for CeHsc70-Δ384 (▾), CeHsc70-Δ512 (▴), CeHsc70-Δ545 (•) and CeHsc70 (▪) were fit to single exponential functions. The inset shows the initial phase of the hydrolysis reaction within the first 200 s.