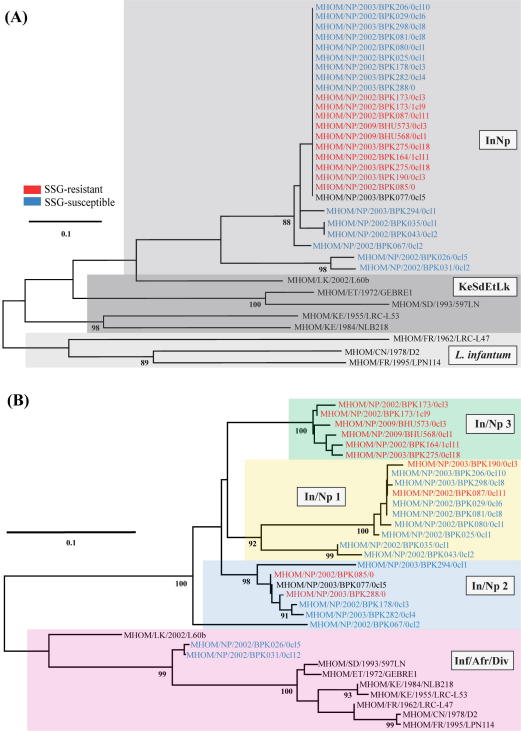
Fig. 2.
Phylogenetic relationships of strains from the L. donovani complex from (A) microsatellite and (B) SNP data. Midpoint-rooted neighbour-joining trees constructed for 33 L. donovani complex strains with MEGA using: (A) microsatellite data for K = 3 geographical populations called InNp (grey containing Indian – denoted BHU – as well as Nepalese strains named BPK), KeSdEtLk (dark grey, L. donovani from Kenya, Sudan, Ethiopia and Sri Lanka) and L. infantum (pale grey from China and France) – notably, BPK026/0cl5 and BPK031/0cl12 cluster in the InNp group for microsatellites; and (B) SNP data for K = 4 populations denoted In/Np 1 (yellow), 2 (blue) and 3 (green), which were composed of strains from India and Nepal; and Inf/Afr/Div (mauve), which represented Kenyan, Sudanese, Ethiopian, Sri Lankan, Chinese and French L. donovani complex strains in addition to two Nepalese samples (BPK026/0cl5 and BPK031/0cl12). Populations were determined using structure for both marker types. In vitro responses to SSG are shown as resistant (red) or sensitive (blue). Bootstrap values were determined for 104 (SNPs) or 103 (microsatellites) replicates – only those with more than 87% confidence are shown at each node. Branch lengths are proportional to the genetic distance.