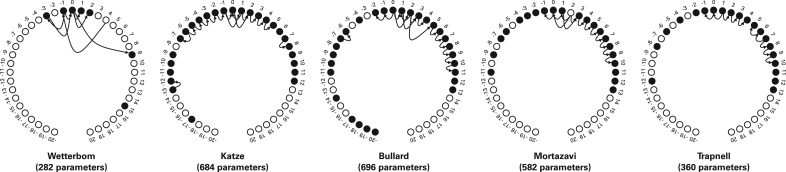
Fig. 3.
The network structures learned on each of the datasets are displayed. Positions are relative to the read start, which is labeled 0. Hollow circles indicate positions that were not included in the model, being deemed uninformative, given the other positions and edges. The number of parameters needed to specify each model is listed in parenthesis below. Applied to data with less bias, a sparser model is trained, as evinced by the Wetterbom dataset. Note that dependencies (i.e. arrows) tend to span a short distances, and nodes tend to have a small in-degree (i.e. have few inward arrows). In practice, we save time in training by prohibiting very distant dependencies (>10, by default) or very high in-degrees (> 4, by default).