Fig. 1.
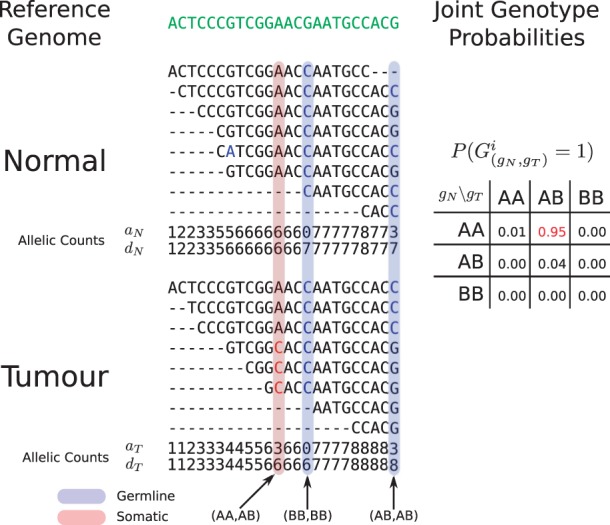
Hypothetical example of the JointSNVMix analysis process. Reads are first aligned to the reference genome (green). Next the allelic counts, which are the number of matches and depth of reads at each position are tabulated. Allelic count information can then be used to identify germline (blue) and somatic positions (red). At the bottom of the Figure, we show the hypothetical probabilities of the nine joint genotypes based on the count data for the somatic position (AA, AB).
