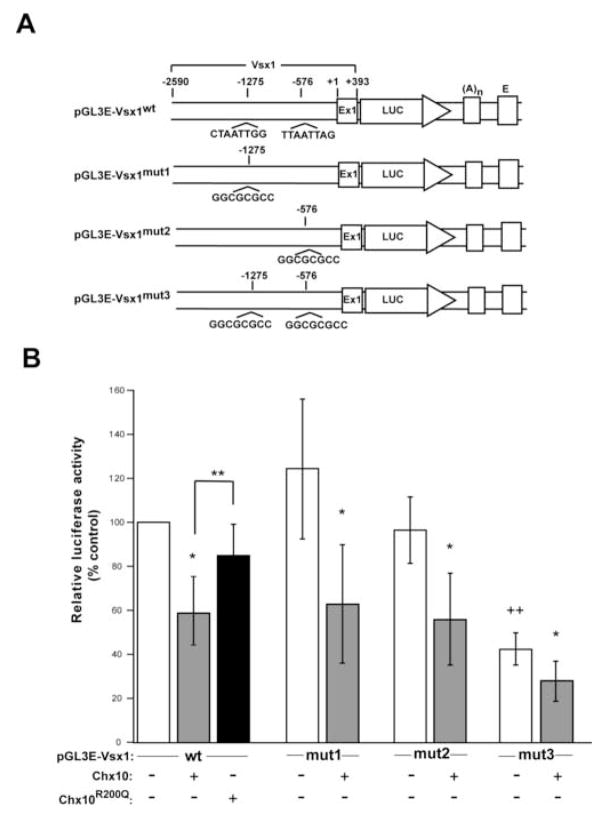
Figure 3. Chx10 expression inhibits luciferase activity in a DNA-binding dependent manner and Chx10 binding sites near the Vsx1 promoter are required for high luciferase activity in HEK293 cells.
(A) Schematic diagrams of reporter constructs with Vsx1 promoters that contain both Chx10 binding sites (pGL3E-Vsx1wt) or with site specific mutations in these sites (pGL3E-Vsx1mut1, mutated at -1275; pGL3E-Vsx1mut2, mutated at −576; pGL3E-Vsx1mut3, mutated at both sites). Region bracketed corresponds to Vsx1 genomic DNA and numbering is relative to predicted Vsx1 transcription start site. (B) HEK293 cells were transfected with each reporter construct, pRL-TK to monitor transfection efficiency, and either an empty expression vector (pCMV; white bars), pCMV-Chx10 (gray bars), or pCMV-Chx10R200Q (black bar). Relative luciferase activity is normalized to the pGL3E-Vsx1wt co-transfected with empty expression vector (the first white bar) and is arbitrarily assigned as 100% activity. The standard deviation for each test condition is shown. Statistical significances (p-values) were tested by two-way repeated measures ANOVA and Tukey tests for multiple comparisons with values transformed by log2. *p<0.001 (p-value of each gray bar compared to each respective white bar); **p =0.0001; ++ p <0.001 (p-value of pGL3E-Vsx1mut3 (last white bar) compared to other reporter constructs (white bars)). Abbreviations: Ex1, exon 1 of Vsx1; LUC, luciferase cDNA; (A)n, SV40 late poly(A) signal; E, SV40 enhancer.