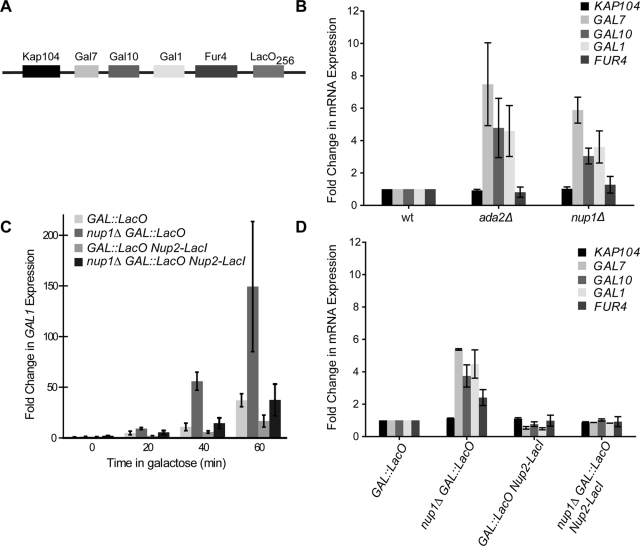
FIGURE 3:
The nuclear periphery specifically regulates expression of galactose-induced genes. (A) Schematic of the GAL locus and adjacent genes on chromosome II, including the integration site of 256 copies of the LacO repeats. (B) mRNA expression levels of GAL locus genes (GAL1, GAL7, GAL10) and neighboring genes (FUR4 and KAP104) in yeast lacking the gene–periphery tether. Wild-type, ada2Δ, or nup1Δ mutant cells were grown in YPD medium to mid-log phase and shifted to YPGal medium for 1 h. mRNA levels were measured by qRT-PCR, and the expression of each gene was normalized to the control gene, TFC1. Fold change of mRNA levels for all genes in ada2Δ or nup1Δ strains was calculated relative to expression in the wild-type strain, defined as 1.0. (C) Kinetics of GAL1 expression with constitutive peripheral tethering. GAL1 gene expression, measured by qRT-PCR in wild-type and nup1Δ strains containing the LacO repeats integrated near the GAL locus with and without the Nup2-LacI gene tether. Cells were grown in YPD medium, and GAL1 expression was induced with 2% galactose for the indicated times. GAL1 mRNA levels were normalized to levels of the control gene TFC1, and the fold change in expression was calculated relative to the baseline expression at the zero time point for each strain. (D) mRNA expression levels of GAL locus genes and neighboring genes with the Nup2-LacI gene tether. mRNA levels were detected by qRT-PCR following 1 h induction with galactose. The expression of each gene was normalized to the control gene TFC1, and the fold change of mRNA levels for all genes was calculated relative to expression in the GAL::LacO strain, defined as 1.0. Error bars represent SEM for three independent experiments.