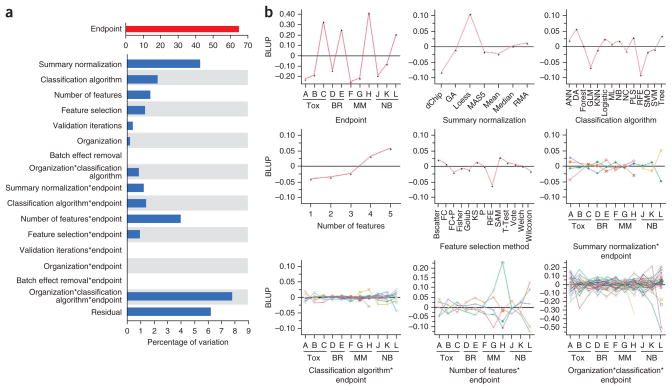
Figure 5.
Effect of modeling factors on estimates of model performance. (a) Random-effect models of external validation performance (MCC) were developed to estimate a distinct variance component for each modeling factor and several selected interactions. The estimated variance components were then divided by their total in order to compare the proportion of variability explained by each modeling factor. The endpoint code contributes the most to the variability in external validation performance. (b) The BLUP plots of the corresponding factors having proportion of variation larger than 1% in a. Endpoint abbreviations (Tox., preclinical toxicity; BR, breast cancer; MM, multiple myeloma; NB, neuroblastoma). Endpoints H and L are the sex of the patient. Summary normalization abbreviations (GA, genetic algorithm; RMA, robust multichip analysis). Classification algorithm abbreviations (ANN, artificial neural network; DA, discriminant analysis; Forest, random forest; GLM, generalized linear model; KNN, K-nearest neighbors; Logistic, logistic regression; ML, maximum likelihood; NB, Naïve Bayes; NC, nearest centroid; PLS, partial least squares; RFE, recursive feature elimination; SMO, sequential minimal optimization; SVM, support vector machine; Tree, decision tree). Feature selection method abbreviations (Bscatter, between-class scatter; FC, fold change; KS, Kolmogorov-Smirnov algorithm; SAM, significance analysis of microarrays).