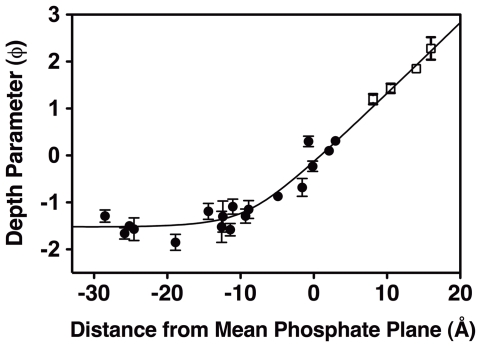
Figure 5. Hyperbolic relationship between spin label depth parameters and membrane penetration depths in the optimized, self-consistent EPR docking model.
As described in Methods, the crystal structure of the GRP1 PH domain co-complex with IP4 (1FGY [22]) was modeled with MTSSL spin labels at the 18 chosen positions, then docked to the target bilayer using an interactive procedure that optimizes the known hyperbolic relationship between the measured spin label EPR depth parameters and the calculated spin label membrane penetration depths. Shown are the measured depth parameters for the protein spin labels (filled symbols) and the calibration lipid spin labels (open symbols), as well as the calculated membrane depth for each spin label in the final optimized, self-consistent EPR membrane docking model (Figure 6). The excellent agreement with the best-fit hyperbola (solid curve) emphasizes the high quality of the docking model. Depth parameters were measured by EPR power saturation (Methods) at 23°C and samples contained 10–200 µM protein, 40 mM total lipid as SUVs, 25 mM HEPES, 140 mM KCl, 15 mM NaCl, 0.5 mM MgCl2, pH 7.4. Except where otherwise indicated, errors are propagated from the errors of the accessibility parameters (Π(NiEDDA) and Π(O2)) used to calculate the depth parameter (Eq. 1), n≥15 power settings were used for each accessibility parameter measurement.