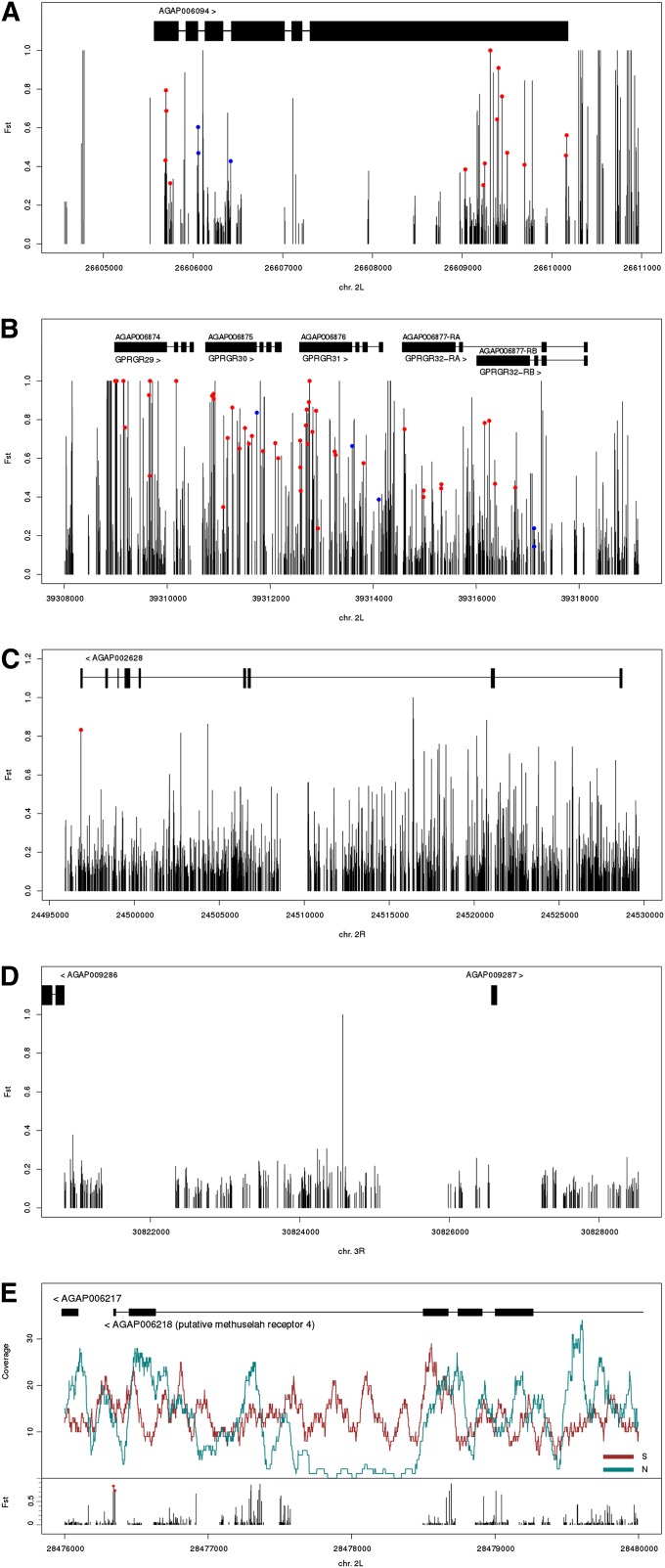
Figure 7 .
Elevated FST or copy-number variation between northern and southern Cameroon populations in protein coding genes. FST is plotted for individual SNPs along the gene models (solid rectangles, exons; horizontal lines, introns). Nonsynonymous and splice site mutations are indicated by red and blue circles, respectively. Divergence in (A) the JIL-1 ortholog, AGAP006094 in the 2La rearrangement; (B) the cluster of gustatory receptors GPRGR29-32 in the 2La rearrangement; (C) AGAP002628 in the 2Rb rearrangement, a putative member of the immunoglobulin superfamily; (D) an intergenic region upstream and nearby two genes in a collinear region on 3R; (E) the Methuselah receptor 4 ortholog AGAP006218 in the 2La rearrangement. In addition to FST at individual SNPs, read coverage is shown for each population (northern, green line; southern, red line) in relation to the gene model.