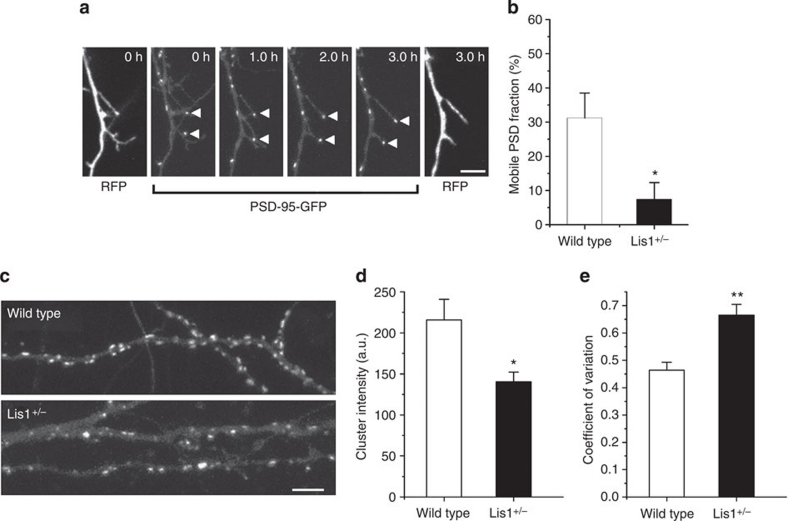
Figure 7. Effect of manipulating the Lis1 gene in cortical slices.
(a) Time-lapse images of an interneuron maintained in cortical slices from a Lis1hc/+ mouse and transfected with expression plasmids encoding Cre recombinase and PSD-95-GFP, together with a Cre indicator plasmid. Arrowheads indicate stationary PSD puncta within dendritic protrusions. Scale bar, 5 μm. (b) Fraction of mobile PSD-95-GFP puncta in dendritic protrusions of control and Lis1−/+ interneurons (31.2±7.3% for wild type and 7.4±4.9% for Lis1−/+, n=12 cells in nine slice cultures for wild type, n=9 cells in nine slice cultures for Lis1−/+). Significant differences (unpaired Student's t-test): *P<0.05. (c) Distribution and morphology of PSD-95-GFP puncta in cortical interneurons from wild-type or Lis1hc/+ mice and transfected with expression plasmids encoding Cre recombinase and PSD-95-GFP, together with a Cre indicator plasmid. Scale bar, 5 μm. (d) Total fluorescence of individual PSD-95-GFP puncta was lower in interneurons of the Lis1−/+ genotype than in wild-type interneurons (Lis1−/+: 140±12 a.u., n=28 cells in seven slice cultures; wild type: 215±25 a.u., n=27 cells from seven slice cultures). Significant differences (unpaired Student's t-test): *P<0.05. (e) Larger variation in the total fluorescence of PSD-95 puncta in interneurons carrying the Lis1−/+ genotype. The average coefficient of variation for the total fluorescence of PSD-95 puncta in each dendrite was significantly larger in Lis1−/+ interneurons (Lis1−/+: 0.66±0.039, n=27 cells from seven slice cultures; wild type: 0.46±0.029, n=28 cells from seven slice cultures). Significant differences (unpaired Student's t-test): **P<0.01. All numeric data are mean±s.e.m.