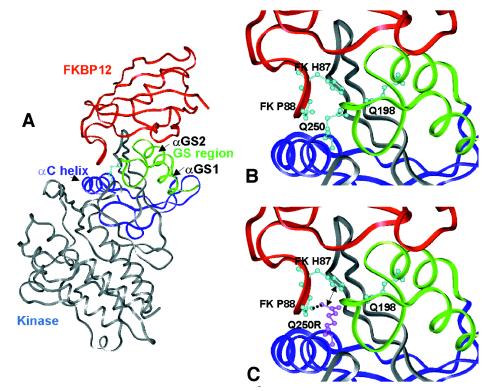
Figure 5.
Modeling of the effects of Q250R substitution on the human TβR-I/FKBP12 complex structure. (A) Experimental structure of TβR-I in complex with FKBP12 (Protein Data Bank identifier 1B6C). Q250, located at the C terminus of αC helix, is shown in a ball-and-stick representation. The ovine BMPR-IB model is very similar to TβR-I (0.1 Å rms between the two structures: 326 superimposable Cα). (B) Q250 is sandwiched between the FKBP12 flap (one side of the FKBP12 active site in which helix αGS2 is embedded) and the αGS1/αGS2 loop (GS region), which undergoes phosphorylation after kinase activation. The movement of αC helix, allowed by the GS region phosphorylation and/or FKBP12 dissociation, is likely to be responsible partly for kinase activation (23). Q250 does not form any obvious bond with the GS region nor with FKBP12 (Q198 Oɛ1 atom, the nearest neighbor, is 3.5 Å distant from Q250 Nɛ2 atom). (C) Consequence of the Q250R mutation. In the most probable conformer, R250 is predicted to form a strong hydrogen bond through its Nη1 atom with the main chain oxygen of FKBP12 P88 (dashed line). This interaction between FKBP12 and the receptor should be reinforced by a nearly parallel stacking interaction between π electrons of R250 and FKBP12 H87 (e.g., 3.2 Å between R250 Nɛ and FKBP12 H87 Nɛ2; see arrow). The same prediction can be made for the ovine BMPR-IB model that has been fitted on the TβR-I/FKBP12 complex.