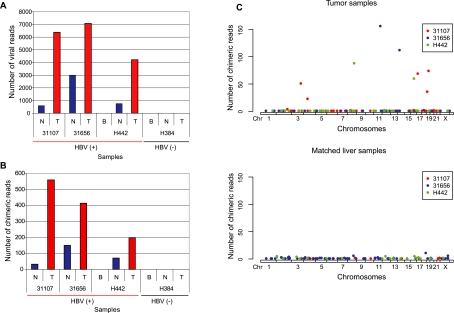
Figure 1.
Tumor-specific clonal expansion of virus-integrated hepatocytes in HCC. For each sequenced human genome, viral integration is quantified as the total number of paired-end reads where at least one arm maps to the HBV genome (A), the number of human-viral chimeric reads (B), and the number of chimeric reads as a function of the genomic location of HBV integration sites in the human genome (C). In contrast to the matched nontumor samples, tumor samples carry a few loci with a substantially larger number of chimeric reads. (T) Hepatocellular carcinoma tumor samples. (N) Matched nontumor liver samples. (B) Blood samples. The legend in panel C indicates internal identifiers for the three HBV positive patients in this study.