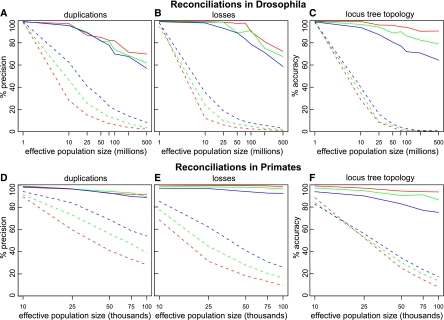
Figure 5.
Increased performance of DLCoalRecon in simulated fly and primate gene trees. DLCoalRecon (solid) and MPR (dashed) were used to reconcile 500 fly and 500 primate simulated gene trees. Duplications and losses were simulated at rates that were the same as (1×, red), twice (2×, green), and four times (4×, blue) the rate estimated in real data. Increased performance is seen both in the precision of inferring duplications and losses (A,B,D,E) as well as the accuracy of reconstructing the locus tree topology (C,F).