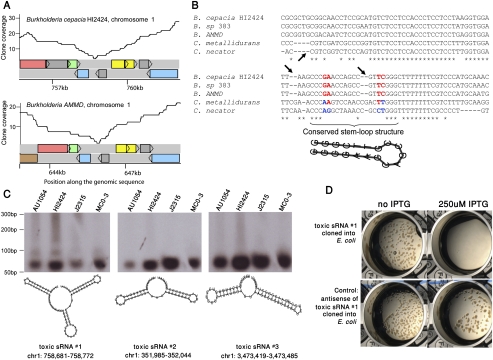
Figure 2.
Toxic small RNAs reside in intergenic gaps. (A) Reproducibility of cloning deficiency of an intergenic region in Burkholderia. Coverage plots of a syntenic 6-kb region in chromosome 1 of two Burkholderia species. Colored rectangles represent genes. This region harbors toxic small RNA #1 (tsRNA #1). (B) Multiple alignment of the cloning-deficient intergenic region. The orthologous region to those shown in panel A was aligned between five closely related organisms. Arrows depict deletions that are not divisible by 3, indicating that this region does not code for protein. Compensatory, stem-preserving mutations are colored red/blue, indicative of conserved RNA structure. (C) Northern blot analysis of three tsRNAs found in intergenic gaps in Burkholderia. Radio-labeled probes for each of the tsRNAs were hybridized to the total RNA of four Burkholderia species: Burkholderia cenocepacia AU1054, B. cenocepacia HI2424, B. sp. J2315, and B. sp. MC0-3. Predicted secondary structures of small RNAs were calculated using RNAfold (Hofacker et al. 1994). Coordinates are given relative to the B. cenocepacia HI2424 genome. Two bands of tsRNA #1 probably indicate post-transcriptional sRNA processing of the long form (94 bases) into a shorter form (∼60 bases). (D) Toxicity of tsRNA #1 in E. coli. tsRNA #1 from B. cenocepacia HI2424 was cloned into E. coli under the control of IPTG-inducible promoter. (Top left) Colonies of E. coli grow when the expression of the tsRNA is suppressed. (Top right) Growth inhibition is observed following activation of tsRNA expression. (Bottom) Growth inhibition is not observed when the antisense of the small RNA is expressed in the same system.