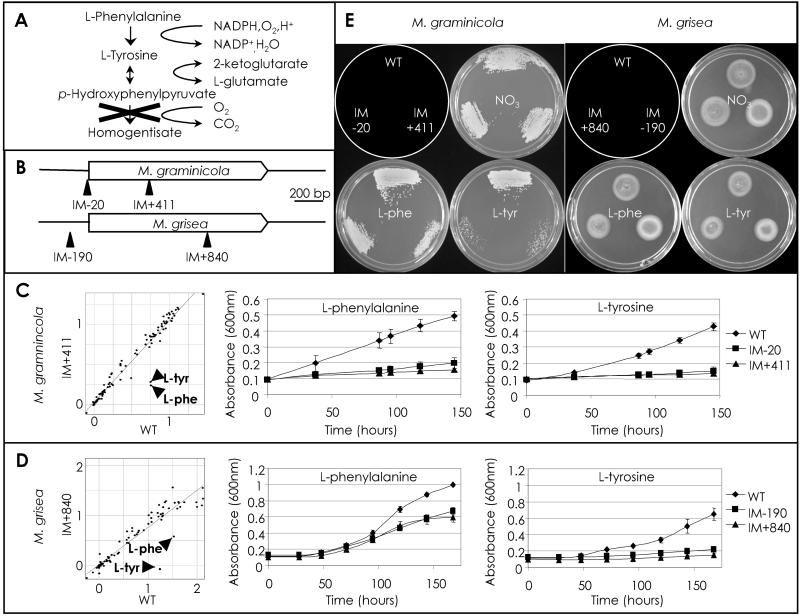
Figure 5.
Analysis of M. graminicola and M. grisea HPD4 TAGKO mutants. (A) The l-phenylalanine (l-phe) and l-tyrosine (l-tyr) degradation pathway. X indicates the pathway block by inactivation of HPD4. (B) M. graminicola and M. grisea HPD4 alleles showing the location of the TAGKO insertions. (C) Graphs showing growth of M. graminicola WT vs. IM+411 at 7 days in the presence of 95 different nitrogen sources (l-Phe and l-Tyr are indicated) and time courses of M. graminicola WT, IM-20, and IM+411 in MM-N + Phe and MM-N + Tyr, respectively. (D) Graphs showing growth of M. grisea WT vs. IM+840 at 7 days in the presence of 95 different nitrogen sources (l-Phe and l-Tyr are indicated) and time courses of M. grisea WT, IM-190, and IM+840 in MM-N + Phe and MM-N + Tyr, respectively. (E) Plate images showing growth recovery on complete medium of M. graminicola WT, IM-20, and IM+411 (after 8 days) and M. grisea WT, IM-190, and IM+840 (after 5 days) after incubation in MM, MM-N + Phe or MM-N + Tyr, as indicated. The schematic plate diagram depicts the location of each strain.