Abstract
Annulatascus nilensis sp. nov., from freshwater habitats in Egypt, is described, illustrated and compared to other species in the genus. Phylogenetic analyses of its LSU rDNA sequence with similar fungi placed the new species in the genus Annulatascus (Annulatascaceae, Sordariomycetidae incertae sedis). Annulatascus nilensis is characterized by immersed ascomata with an ascomatal neck oriented horizontally to the substrate surface, asci with a long, narrow stalk and massive bipartite apical ring, and 5–11-septate, hyaline ascospores surrounded by a large irregular, granular sheath that is not seen in water.
Keywords: Ascomycota, molecular phylogeny, Phragmites, Sordariomycetes, subtropics
INTRODUCTION
The generic name Annulatascus was introduced by Hyde (1992) to accommodate two freshwater fungi, A. velatisporus (the type species) and A. bipolaris, which were collected from submerged wood in Australia. Fifteen species are currently included in the genus (Barbosa et al. 2008), most of which were described from freshwater habitats in the tropics (Barbosa et al. 2008, Shearer et al. 2010, <www.fungi.life.uiuc.edu>). However, two of the species were discovered on palm rachides on land (Fröhlich & Hyde 2000). Annulatascus species are characterized by dark, immersed or superficial perithecioid ascomata; wide, tapering paraphyses; long cylindrical asci with a relatively massive refractive bipartite apical ring; and hyaline, fusiform, sheathed or appendaged ascospores. During an ongoing investigation of freshwater fungi on wood submerged in the River Nile in Egypt, we discovered a previously unknown species of Annulatascus which is described and illustrated here, and compared to other species in the genus using morphological and molecular data. Phylogenetic analyses of the LSU rDNA sequence of the new fungus placed the new fungus in the same clade as the type species of the genus ( Annulatascus velatisporus) and A. hongkongensis.
MATERIALS AND METHODS
Collection
Submerged decayed wood was recovered from the River Nile in the Sohag governorate. Samples were placed in clean plastic bags, and taken to the laboratory where they were examined immediately under a stereomicroscope for fungal spore-producing structures. Samples were then incubated on moist filter paper in sterile plastic boxes, and examined periodically over a 6 mo incubation period. Single-ascospore cultures of the new fungus were obtained by cutting open ascomata with a sterile razor-blade, and the centrum tissue containing ascospores was removed with sterile forceps and placed in sterile freshwater. Small drops of the ascospore suspension were placed on Czapek’s medium in Petri dishes and incubated at 22 °C in the dark. Germinated ascospores were transferred to new Czapek’s medium Petri dishes with sterile forceps and incubated at 22 °C in the dark. Photographs were taken using an Olympus B×51 differential interference contrast light microscope and Olympus DP12 digital imaging system (Olympus Corporation, Tokyo). Reference specimens were prepared by drying at 60 °C for 24 h, and deposited along with fungal cultures in the collections of the Department of Botany, Faculty of Science, Sohag University, Egypt. Representative slides and a portion of the material selected to serve as the holotype were deposited in the Biosystematics Reference Collection of CAB International (IMI). Pure cultures of the new fungus used in this study, were deposited at the Extremobiosphere Research Center, Japan Agency for Marine-Earth Science and Technology (JAMSTEC), under accession number “MF808”.
DNA extraction, sequencing, and phylogenetic analysis
DNA was extracted from pure fungal mycelium using the Microbial DNA Extraction Kit (MOBIO; Mo Bio Laboratories, Carlsbad, CA) according to the manufacturer’s instructions. Partial LSU ribosomal DNA was amplified using primers LR0R and LR7 (Bunyard et al. 1994). PCR reactions, cycling parameters and sequencing were carried out as described by Abdel-Wahab et al. (2009). Sequences were assembled using Sequencher v. 4.2.2 (Gene Codes Corporation). Sequences were aligned with pertinent ones retrieved from GenBank using Clustal X (Thompson et al. 1997) and optimized manually. The positions where one or more species contained a length mutation or ambiguously aligned regions were not included in the phylogenetic analysis. Nucleotide sequence phylogenies were constructed using PAUP v. 4.0b10 (Swofford 2002). Maximum-likelihood (ML) analyses (Felsenstein 1981) were performed using heuristic searches with the random stepwise addition of 100 replicates and tree bisection-reconnection (TBR) rearrangements in effect. The optimal model of nucleotide substitution for the ML analyses was determined using hierarchical likelihood ratio tests as implemented in Modeltest v. 3.7 (Posada & Crandall 1998). The model selected as the best fit for LSU rDNA data set was TrN+G. For the bootstrap analyses (Felsenstein 1985), 100 replicates were generated with five random additions and TBR. Maximum-parsimony (MP) trees were obtained by 100 random addition heuristic search replicates using PAUP, and 1 000 bootstrap replicates were performed employing five random addition heuristic searches. Posteriori probability values were obtained using MrBayes v. 3.1.2 (Huelsenbeck & Ronquist 2001, Ronquist & Huelsenbeck 2003) with the GTR+G model that was determined using MrModeltest v. 2.2 (Nylander 2004). Nodal supports in the Bayesian analyses were examined by posterior probabilities from five million generations that were run in four chains with sampling every 100 generations, yielding 50 000 trees, of which the first 12 500 were discarded as “burn in”. The LSU sequence of the ex-type isolate used in this study was deposited at GenBank under the accession number “HQ616536” (MF 808, JAMSTEC). The alignment was deposited in TreeBASE (<treebase.org>) under accession number S10747, and the description of the new species was deposited in MycoBank (<mycobank.org>; Crous et al. 2004).
RESULTS
Phylogenetic analyses
The partial LSU rDNA sequence of Annulatascus nilensis aligned with representatives of the Annulatascaceae, along with representatives of Halosphaeriales, Sordariales, Trichosphaeriales, and Xylariales. In total, the LSU rDNA dataset included 40 taxa of which three belonged to Pezizales were used as outgroup. The dataset consisted of 879 characters, of which 329 were constant, 183 were variable but parsimony-uninformative, and 367 were parsimony informative. The ten most parsimonious trees produced using heuristic search parameters were of equal length with 1 308 steps, and have a CI of 0.4602, an RI of 0.5864, and a Rescaled CI of 0.2699. The maximum likelihood analysis produced one tree with a likelihood score of 6995.34. Bayesian analyses yielded two trees, of which one is shown here as Fig. 1. The maximum parsimony and likelihood analyses both produced trees with similar topologies (data not shown).
Fig. 1.
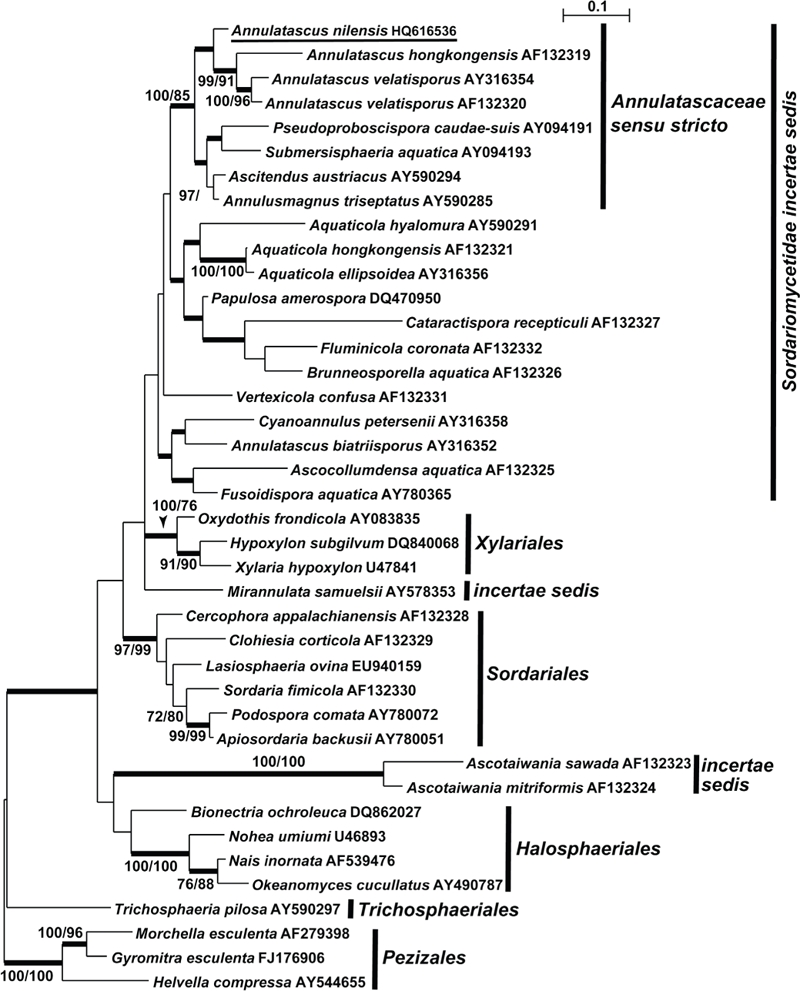
Phylogenetic relationships of Annulatascus nilensis and similar fungi, based on the nucleotide sequences of LSU rDNA. The Bayesian phylogenetic tree (MrB) was constructed as described in the text. The numbers indicate pp values ≥ 95 % (in bold), MP, ML bootstrap values ≥ 70. The new species, Annulatascus nilensis, is underlined.
Annulatascus nilensis was found to be phylogenetically close to A. velatisporus (the type species of Annulatascus) and A. hongkongensis, and was nested within a highly supported clade (100/100/85 for Bayesian/MP/ML respectively) which included the genera Annulatascus, Annulusmagnus, Ascitendus, Pseudoproboscispora, and Submersisphaeria. This clade, was phylogenetically distant from clades of other Annulatascaceae genera, and represents the family Annulatascaceae s. str. (Fig. 1). Phylogenetic analyses of LSU rDNA showed Annulatascus as currently circumscribed to be polyphyletic, with A. biatriisporus phylogenetically distant from A. velatisporus (Raja et al. 2003, Campbell & Shearer 2004).
Taxonomy
Annulatascus nilensis Abdel-Wahab and Abdel-Aziz, sp. nov.
MycoBank MB517837
Fig. 2.
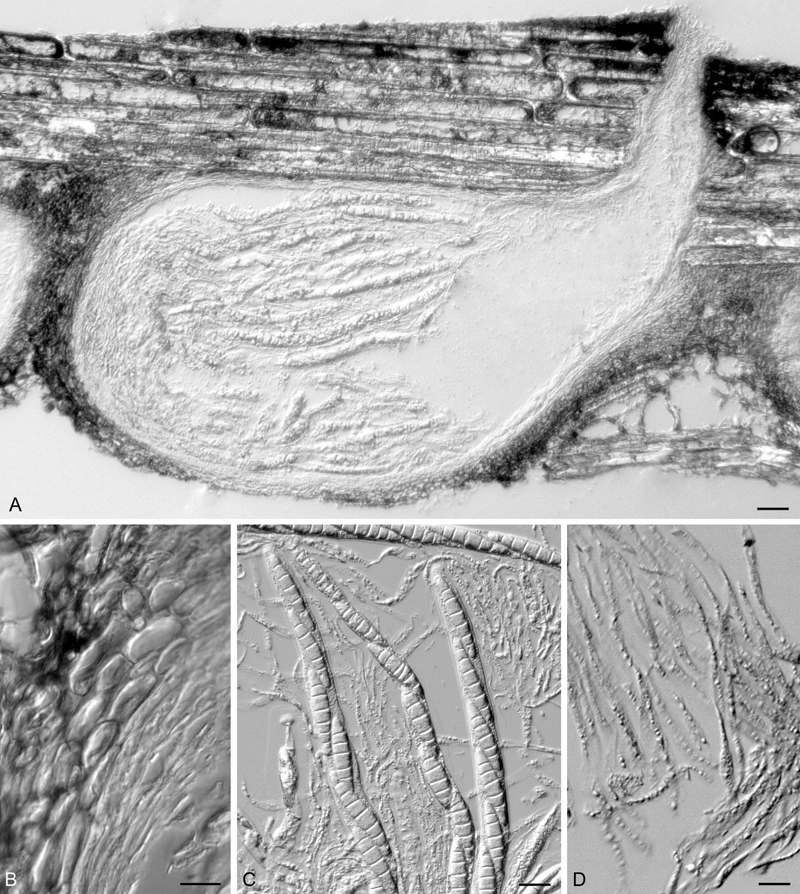
Annulatascus nilensis (holotype). A. Vertical section through ascomata showing the hymenium at the base of the ascomatal venter and periphysate neck. B. Section through the peridial wall. C. Squash of the ascoma venter showing asci and paraphses. D. Paraphyses. Bars: A = 20 μm, B = 5 μm, C, D = 10 μm.
Fig. 3.
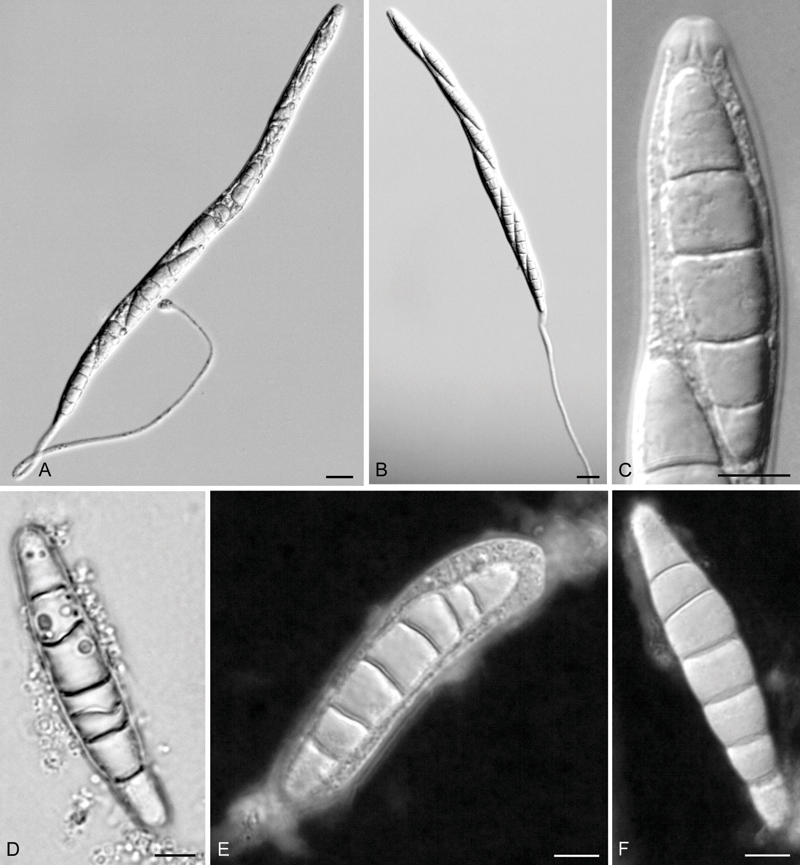
Annulatascus nilensis (holotype). A, B. Asci at different stages of maturity; note the long narrow stalk. C. Ascus with apical mechanism. D–F. Ascospores surrounded by an irregular, granular sheath that peels off at a later stage. Bars: A, B = 10 μm, C–F = 5 μm.
Etymology: From the Latin word Nilus, the River Nile, where the fungus was collected.
Ascomatibus immersis, obyriformibus, axe principali horizontali, nigris, coriaceis, ostiolatis, papillatis, solitariis vel gregariis. Collis assurgentibus, periphysatis. Paraphysibus raro septatis, hyalines, apice attenuato. Ascis 260–400 × 12–14 μm, octosporis, cylindricis, unitunicatis, cum apparato apicali. Ascosporis uniserialibus vel imbricate uniserialibus, 32–52 × 7–10 μm, fusiformibus, 5–9(–11)-septatis, ad septa constrictus, hyalinis, laevibus, cum vagina ampla iregulari granulose.
Typus: Egypt: Sohag, in the River Nile, on decayed submerged stems of Phragmites australis, Feb. 2006, A.E. Abdel-Aziz (IMI 397966 – holotypus; culture ex-type MF 808; GenBank accession no. HQ616536).
Ascomata 400–600 μm long, 220–280 μm wide, immersed, obyriform, oriented horizontally to the substrate surface, black, coriaceous, ostiolate, papillate, solitary or gregarious (Fig. 2A). Peridium 20–26 μm thick, forming a textura angularis of thick-walled flattened cells, the cells dark brown and encrusted with pigmented particles in the outer parts, and hyaline and unencrusted towards the inside (Fig. 2A, B). Ostiolar necks curved upward, 240–360 μm long, 96–112 μm wide. Hamathecium of periphyses lining the ostiolar canal, to 20 μm long and 1.5–2 μm wide, and paraphyses in the centrum which are 3–12 μm wide at the widest part, rarely septate, unbranched, hyaline, and taper distally (Fig. 2C, D). Asci 260–400 × 12–14 μm (av. = 322.5 × 12.2 μm, n = 20), cylindrical, unitunicate, deciduous in water mount, persistent, with a tapered elongated base when mounted in water. and a large non-amyloid, refractive, bipartite apical ring 3–4 μm high × 5–6 μm wide (Fig. 3A–C), discharge by deliquescence, 8-spored. Ascospores 32–52 × 7–10 μm (av. = 40.8 × 8 μm, n = 41), uniseriate to overlapping uniseriate, fusoid, 5–9(–11)-septate, constricted at the septa, hyaline, smooth-walled, surrounded by a large, irregular, granular sheath (Fig. 3D–F) that is not evident in water mounts.
Notes: Annulatascus nilensis has ascospore dimensions that overlap with those of A. biatriisporus, A. velatisporus, and A. tropicalis. However, the first two species have aseptate ascospores, while A. tropicalis has 1–3-septate ascospores that also lack a mucilaginous sheath or appendages (Hyde 1992, Tsui et al. 2002). Annulatascus nilensis differs from A. fusiformis in that the later has ascospores that are smaller, 1–5-septate, thick-walled, straight or slightly curved, with a warty ornamentation in the apical and subapical regions (TEM), bipolar mucilaginous pad-like apical appendages, and ascomata with a longer neck (Hyde & Wong 2000). Annulatascus nilensis differs from all the described Annulatascus species in having asci with an elongated narrow stalk (when mounted in water) and 5–11-septate ascospores surrounded by a large, irregular, granular sheath. An elongating basal stalk is also found in Cataractispora receptaculorum, but the ascospores of that species are smaller, more tapered, with polar pads that unfurl in water into long thin filamentous appendages, and are not surrounded by a gelatinous sheath (Ho et al. 2004).
Acknowledgments
M.A. A.-W. is grateful to Takahiko Nagahama for his support and guidance during this work, and to JSPS for the award of a postdoctoral fellowship. We are very grateful to Uwe Braun (Martin-Luther Universität, Halle/Wittenburg) for revising the Latin diagnosis. This work was funded by grants from the Japan Society for the Promotion of Science (JSPS) (Nos.185701000001 and 18-06620). Sohag University is acknowledged for providing funds for collecting samples. M.A. A.-W. is also grateful to the Third World Academy of Science (TWAS) for the award of a research grant (no. 03-117 RG/BIO/AF/AC).
REFERENCES
- Abdel-Wahab MA, Nagahama T, Abdel-Aziz FA. (2009) Two new Corollospora species and one new anamorph based on morphological and molecular data. Mycoscience 50: 147–155 [Google Scholar]
- Barbosa FR, Gusmão LFP, Raja HA, Shearer CA. (2008) Annulatascus apiculatus sp. nov., a new freshwater ascomycete from the semi-arid Caatinga biome of Brazil. Mycotaxon 106: 403–407 [Google Scholar]
- Bunyard BA, Nicholson MS, Royse DJ. (1994) A systematic assessment of Morchella using RFLP analysis of the 28S rRNA gene. Mycologia 86: 762–772 [Google Scholar]
- Campbell J, Shearer CA. (2004) Annulusmagnus and Ascitendus, two new genera in the Annulatascaceae. Mycologia 96: 822–833 [DOI] [PubMed] [Google Scholar]
- Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G. (2004) MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22 [Google Scholar]
- Felsenstein J. (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. Journal of Molecular Evolution 17: 368–376 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Fröhlich J, Hyde KD. (2000) Palm microfungi. Fungal Diversity Research Series 3 Hong Kong: [Google Scholar]
- Ho WH, Hyde KD, Hodgkiss IJ. (2004) Cataractispora receptaculorum, a new freshwater ascomycete from Hong Kong. Mycologia 96: 411–417 [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17: 754–755 [DOI] [PubMed] [Google Scholar]
- Hyde KD. (1992) Tropical Australian freshwater fungi II. Annulatascus velatispora gen. et sp. nov., Annulatascus bipolaris sp. nov. and Nais aquaticus sp. nov. Australian Systematic Botany 5: 117–124 [Google Scholar]
- Hyde KD, Wong SW. (2000) Annulatascus fusiformis sp. nov., a new freshwater ascomycete from the Philippines. Mycologia 92: 553–557 [Google Scholar]
- Nylander JAA. (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Center, Uppsala University, Uppsala: [Google Scholar]
- Posada D, Crandall KA. (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14: 817–818 [DOI] [PubMed] [Google Scholar]
- Raja H, Campbell J, Shearer CA. (2003) Freshwater ascomycetes: Cyanoannulus petersenii a new genus and species from submerged wood. Mycotaxon 88: 1–17 [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Shearer CA, Raja HA, Schmidt JP. (2010) Freshwater Ascomycetes and their Anamorphs. <fungi.life.uiuc.edu>.
- Swofford DL. (2002) PAUP* 4.0: Phylogenetic analysis using parsimony (*and other methods). Sinauer, Sunderland, MA: [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui CKM, Ranghoo VM, Hodgkiss IJ, Hyde KD. (2002) Three new species of Annulatascus (Ascomycetes) from Hong Kong freshwater habitats. Mycoscience 43: 383–389 [Google Scholar]


