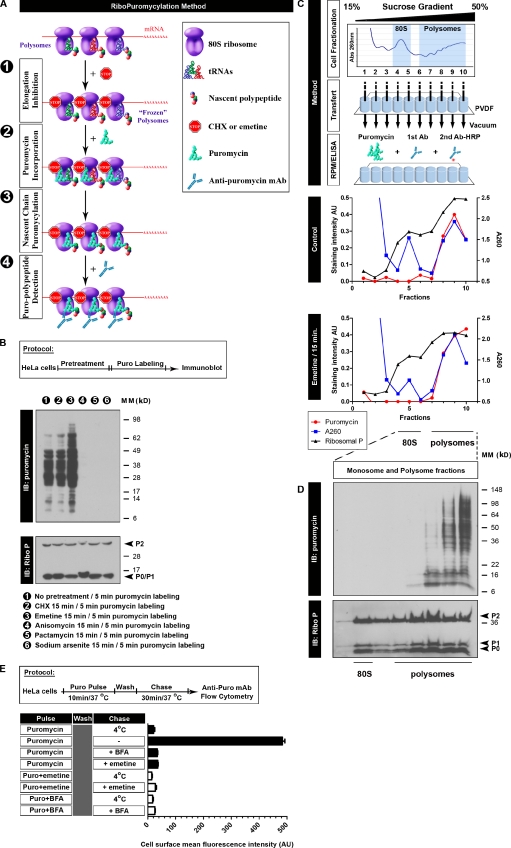
Figure 1.
Characterizing the RPM biochemically. (A) Schematic representation of the RPM. After freezing polysomes with an elongation inhibitor (step 1), PMY is added (step 2) to living cells or subcellular fractions, and nascent chains are puromycylated through ribosome catalysis (step 3). The anti-PMY mAb 12D10 (or other PMY mAbs that we have generated) detects puromycylated nascent chains via immunoblotting or indirect immunofluorescence (step 4). (B) Anti-PMY immunoblotting (IB) of total HeLa cell lysates from cells incubated with PMY for 5 min and other inhibitors as indicated. The smear of proteins represents C-terminally puromycylated nascent chains released from ribosomes. Chain elongation inhibitors do not effect (CHX) or enhance (emetine) nascent chain puromycylation, whereas protein synthesis inhibitors that deplete nascent chains by blocking initiation while allowing chain elongation and completion (pactamycin, direct initiation inhibitor, and arsenite, indirect initiation inhibitor) prevent puromycylation. Anisomycin blocks puromycylation by competing with PMY binding to ribosomes. On the bottom, blotting with anti–ribosomal P (Ribo P) human autoreactive antisera shows that the results cannot be attributed to lane loading discrepancies. (C) HeLa cells incubated or not incubated with emetine for 15 min were lysed and fractionated on 15–50% sucrose gradients. Fractions were bound to PVDF 96-well plates and incubated with PMY, which results in ribosome-catalyzed nascent chain puromycylation. Ribosomes were detected by A260 of fractions or by ELISA for the ribosomal P proteins (here resolved into the three known species) as detected by human autoimmune antibodies, which establishes that monosomes and 60S subunits bind well to PVDF. Puromycylation was detected by ELISA for PMY using 12D10 and clearly demonstrates that monosomes and free 60S subunits do not stably associate with PMY, which requires nascent chain puromycylation. (D) HeLa cells incubated with emetine were lysed and fractionated on 15–50% sucrose gradients. Monosome- and polysome-containing fractions were labeled with PMY on ice, and nascent chains were identified by immunoblotting with 12D10. Only polysomes demonstrate a significant anti-PMY signal, and from the pattern, it is clear that binding is based on nascent chain puromycylation. As expected, the mean size of puromycylated proteins increases with polysome size because, on average, faster sedimenting polysomes possess more ribosomes, translating longer mRNAs. (E) HeLa cells were pulsed with PMY with or without emetine or BFA. Cells were washed and chased with or without emetine/BFA. The control sample (pulse only, labeled 4°C) was kept cold during the chase. Expression of PMY (Puro) on the surface of live cells was determined by flow cytometry using 12D10. In the absence of inhibitors, some puromycylated nascent chains are sufficiently native to be delivered to and expressed on the cell surface with their C-terminal PMY exposed for detection (the basis for the SUnSET assay). Emetine blocks surface expression to the same extent as low temperature or BFA, which completely blocks egress of membrane proteins from the ER. This experiment functionally establishes that emetine prevents release of puromycylated nascent chains from ribosomes, as determined by the proxy population of cell surface proteins detected by the SUnSET method. Error bars represent the standard deviation of triplicate samples. Ab, antibody; AU, arbitrary units; MM, molecular mass.