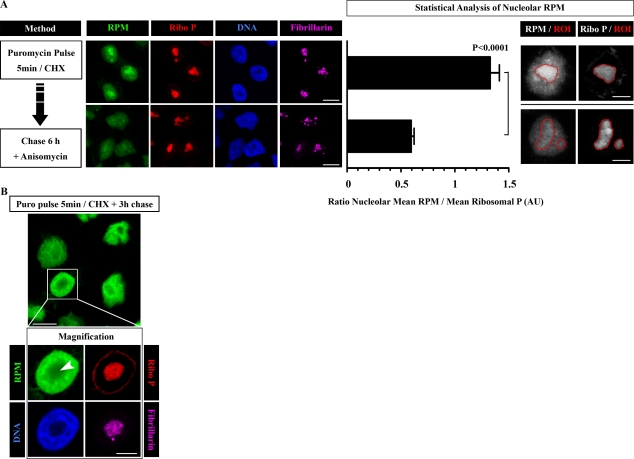
Figure 5.
Nucleolar RPM labeling can be chased. (A) HeLa cells were pulse labeled for 5 min with PMY in the presence of CHX and then chased for 6 h in the presence of anisomycin to prevent further PMY incorporation. For time 0 and time 6 h, cells were RPM stained using NP-40 extraction. Nucleolar RPM staining was measured by defining regions of interest (ROIs) around 15 different nucleoli and calculating the ratio between RPM and ribosomal P (Ribo P) nucleolar mean intensities (mean ± SEM). Statistical analyses, two-tailed unpaired t test. (B) HeLa cells were labeled with PMY CHX for 5 min and then chased for 3 h in the absence of inhibitors. Cells were RPM stained using NP-40 extraction. The arrowhead shows the nucleolus. Bars: (A [left] and B [main image]) 10 µm; (A [right] and B [magnification]) 5 µm. AU, arbitrary unit.