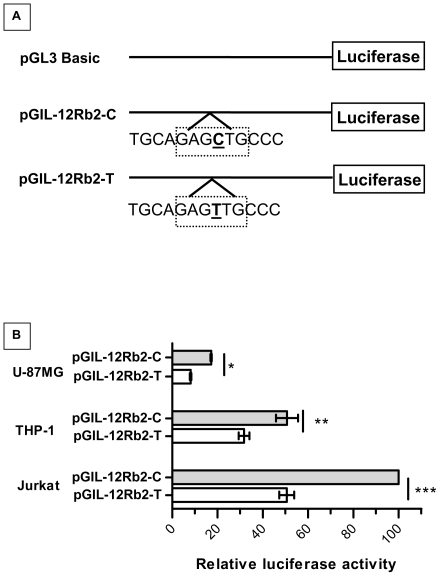
Figure 2. Schematic representation of the vector constructs and the luciferase activity observed in different cell lines.
Panel A. Schematic representation of the promoter constructs, namely pGIL-12Rb2-C (with C allele) and pGIL-12Rb2-T (with T allele) that have been used for the luciferase reporter assays. The region extending from −591 up to +55 of the 5′ region the IL-12Rβ2 promoter was cloned into pGL3-Basic vector. The location of the polymorphic allele at −237 positions has been underlined. The dotted box indicates the AP-4 binding motif in the IL-12Rβ2 promoter. Panel B: The vector constructs pGIL-12Rb2-C, pGIL-12Rb2-T and the pGL3-Basic vector (Negative control) were co-transfected along with pRL-TK vector into U-87, THP-1 and Jurkat cell lines as described in methods. The renilla activity expressed by the pRL-TK vector was used to normalize the transfection efficiency. The reporter gene Firefly luciferase activity was determined for each sample in triplicates, 42 hours post transfection. The relative luciferase activity was expressed as fold increase in the activity compared to the promoter less pGL3-Basic vector and the results has been shown as mean ± SEM of three independent experiments. Student's t test with unequal variances was carried out to compare the transcriptional efficiency (*) of the constructs, (*p = 0.0031; **p = 0.0097, ***p = 0.0001).