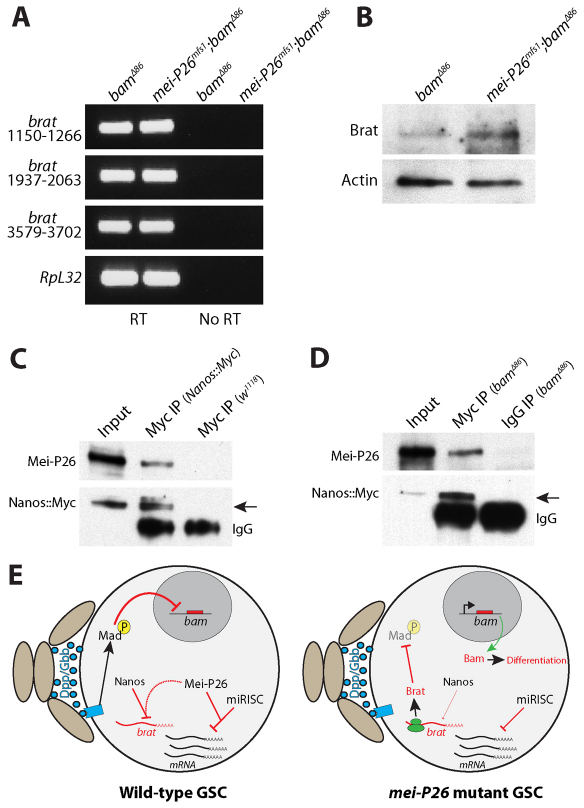
Fig. 7.
Mei-P26 binds to Nanos and regulates Brat protein levels. (A) Ethidium bromide-stained agarose gel showing products from RT-PCR reactions with (RT) and without (No RT) reverse transcriptase. Three different primer pairs specific for different regions of brat mRNA were used to amplify total RNA isolated from bam mutant and mei-P26 bam double-mutant samples. RpL32 was amplified as a loading control. In the RT samples, bam and mei-P26 bam mutants exhibited similar levels of brat mRNA. (B) Western blot analysis of ovarian extracts from bam and mei-P26 bam mutants probed with Brat and Actin antibodies. The mei-P26 bam double-mutant extracts exhibited higher levels of Brat expression than the control extract. (C) Western blot of an anti-Myc co-IP from ovary extract expressing a Myc-tagged nanos transgene. The specificity of the anti-Myc resin was tested using extract from ovaries that did not express the Myc-tagged nanos transgene. (D) Western blot of an anti-Myc co-IP from bamΔ86 mutant extract expressing a Myc-tagged nanos transgene. A rabbit IgG IP of bamΔ86 mutant extract was used as a negative control. co-IP of Mei-P26 with Myc-Nanos was seen in both control ovary and bam mutant ovary extracts (arrows). (E) Model describing the translational regulatory cascades within wild-type and mei-P26 mutant GSCs. Niche cells (light brown ovals) produce Dpp and Gbb, which activate a signal transduction cascade in wild-type GSCs that results in the transcriptional repression of bam. Within wild-type GSCs, Mei-P26 cooperates with both Nanos and miRISC to repress the translation of specific mRNAs. In the absence of Mei-P26, Brat is inappropriately translated resulting in the repression of Mad. Loss of pMAD results in the expression of bam, which causes these mei-P26 mutant GSCs to partially differentiate.