Abstract
Like other Nedd4 ligases, Saccharomyces cerevisiae E3 Rsp5p utilizes adaptor proteins to interact with some substrates. Previous studies have indentified Bul1p and Bul2p as adaptor proteins that facilitate the ligase-substrate interaction. Here, we show the identification of a third member of the Bul family, Bul3p, the product of two adjacent open reading frames separated by a stop codon that undergoes readthrough translation. Combinatorial analysis of BUL gene deletions reveals that they regulate some, but not all, of the cellular pathways known to involve Rsp5p. Surprisingly, we find that Bul proteins can act antagonistically to regulate the same ubiquitin-dependent process, and the nature of this antagonistic activity varies between different substrates. We further show, using in vitro ubiquitination assays, that the Bul proteins have different specificities for WW domains and that the two forms of Bul3p interact differently with Rsp5p, potentially leading to alternate functional outcomes. These data introduce a new level of complexity into the regulatory interactions that take place between Rsp5p and its adaptors and substrates and suggest a more critical role for the Bul family of proteins in controlling adaptor-mediated ubiquitination.
INTRODUCTION
The attachment of ubiquitin to a protein can lead to a wide variety of outcomes, from proteasome degradation to trafficking in the endocytic system (34). The final fate of the ubiquitinated protein appears to be determined by the type of modification that takes place. For example, monoubiquitination and K63-linked polyubiquitination result in endocytosis from the plasma membrane (30), while polyubiquitination through K48 in ubiquitin generally leads to degradation via the 26S proteasome (24). Ubiquitination involves the sequential action of E1, E2, and E3 enzymes, and it is the E3s (or ubiquitin ligases) that control substrate specificity and (with particular E2s) the type of ubiquitin modification that occurs (6, 27). Accessory proteins, such as deubiquitinating proteins (DUBs) and chain-extending ligases (E4s), have also been shown to be important in regulating polyubiquitination (4, 13).
There are two main families of E3s containing either a U-box/RING (really interesting new gene) or HECT (homologous to the E6-associated protein C terminus) domain (7, 16). The RING family members are scaffold proteins that bring together E2 and the target to facilitate ubiquitination (16), while HECT ligases transiently accept ubiquitin from E2 before transferring it to the target protein (7). The Nedd4 (neural precursor cell expressed, developmentally downregulated 4) family members are ubiquitin ligases with a conserved domain architecture consisting of a phospholipid-binding C2 domain, 1 to 4 WW domains (which are protein-protein interaction motifs), and a C-terminal HECT domain (29). There are nine members of the Nedd4 family found in humans, while Saccharomyces cerevisiae contains only one, Rsp5p (29). Nedd4 ligases are involved in regulating endocytosis of receptor proteins and transporters at the plasma membrane (9, 10, 11, 20, 25), sorting of proteins to the yeast vacuole (9, 11), processing of transcription factors into active forms (14, 28), controlling neural development (15), and regulating DNA repair mechanisms (3), and members of the Nedd4 family in humans have been linked with numerous cancers (2).
Generally, the Nedd4 family members interact with targets through their WW domains, which bind short peptide sequences called PY motifs (XPXY) (33). Not all Nedd4 family targets contain PY motifs, and in these cases, adaptor proteins are required to bind both the ligase and the substrate to facilitate ubiquitination (21). Numerous Rsp5p adaptors have been identified in yeast, particularly in relation to the sorting of membrane proteins in the endocytic system (18). Rsp5p adaptors include Bsd2p and Tre1/2p, which are involved in the trafficking of proteins via the multivesicular body (MVB) pathway (11, 32); Ear1p and Ssh4p, two redundant proteins required for the trafficking of numerous cargos originating from the Golgi apparatus and plasma membrane (19); and the nine arrestin-related proteins (Art1p to -9p) required for ubiquitin-mediated endocytosis (20, 25, 26).
Bul1p and Bul2p (binds ubiquitin ligase) are soluble PY motif-containing proteins that bind to and regulate Rsp5p (35, 36). Bul1p and Bul2p have been shown to control the sorting of a number of proteins, including the amino acid permease Gap1p in response to nutrient availability (10), the copper-dependent endocytosis of the transporter Ctr1p (21), and endocytosis of the uracil permease Fur4p (25). In addition, Bul1/2p are required for response to heat and osmotic stress and to DNA damage (3, 35, 36). The roles of Bul1p and Bul2p in the polyubiquitination of Gap1p lead to the suggestion that they may be ubiquitin chain-extending E4 enzymes (10). However, the identification of a small conserved domain and the observation that Bul1p shows some degree of functional redundancy with the arrestin-like proteins led Nikko and Pelham (25) to postulate that Bul1/2p are distantly related arrestin-like adaptor proteins.
Here, we report the exciting identification of a further member of the Bul family, Bul3p, in S. cerevisiae. Like the other Bul proteins, Bul3p is involved in some, but not all, cellular processes that require Rsp5p. Surprisingly, on further analysis of the phenotypes of BUL null mutants, we found the Bul proteins can antagonistically regulate the Rsp5p-mediated sorting of the plasma membrane transporter proteins Smf1p and Can1p. In addition, the phenotypes of the BUL gene deletions suggest complex roles of Rsp5p and the Bul proteins in some cellular pathways, and we postulate that, rather than being adaptors, the Bul proteins may instead allow efficient recruitment and removal of adaptors to modulate the activity of Rsp5p.
MATERIALS AND METHODS
BUL gene deletions.
All the strains used are derivatives of BY4742 (MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0). Single-gene deletions of BUL1 and BUL2 were obtained from the Euroscarf gene knockout collection, and a double deletion was made by mating and sporulation. All BUL3 null mutations were made by replacing the ynr069/ynr068 open reading frames (ORFs) with the Schizosaccharomyces pombe HIS5 marker. The BUL3 ORF was amplified from yeast genomic DNA using PCR, the DNA sequence was obtained (Eurofins, London, United Kingdom), and the protein sequence was compared to those of BUL1 and BUL2 using Sequence Analysis software (Informagen Inc.).
BUL3 complementation and immunoprecipitation.
The BUL3 ORF was inserted into a YCplac33 CEN URA3-derived shuttle vector that adds an N-terminal hemagglutinin (HA) epitope tag under the control of the TPI1 promoter. HA-Bul3p was visualized in equalized total protein extracts from log-phase yeast cultures using immunoblotting with monoclonal anti-HA antibodies (Sigma-Aldrich, Dorset, United Kingdom) and infrared (IR)-dye-conjugated secondary antibodies (Li-Cor Bioscience, Cambridge, United Kingdom). Immunoprecipitation was performed using EZview Red Anti-HA affinity resin (Sigma-Aldrich, Dorset, United Kingdom) according to the manufacturer's protocol using total extracts from log-phase cells, followed by simultaneous immunoblotting with monoclonal anti-HA and polyclonal anti-Nedd4 (Abcam, Cambridge, United Kingdom) primary and IR-dye-conjugated secondary antibodies. For complementation studies, site-directed mutagenesis using a QuikChange protocol (Stratagene, Stockport, United Kingdom) and the HA-Bul3p construct described above as a template was used to remove the BUL3 internal stop codon and to produce a Y100A (ΔPY) construct.
Growth assays.
Yeast cultures were grown overnight in yeast extract-peptone (YEP) agar plus 2% (wt/vol) glucose (YPD), the culture density was equalized according to the A600, and 10-fold dilutions were spotted out onto either YPD agar (or YEP agar plus 2% [vol/vol] ethanol) or synthetic complete agar minus arginine (plus 2% glucose) for canavanine experiments, with the addition of 25 μM CdCl2, 3 μM phleomycin (Melford Laboratories, Ipswich, United Kingdom), 1 mM l-azetidine-2-carboxylic acid (AZC) (Sigma-Aldrich, Dorset, United Kingdom), or 14 μM canavanine (Sigma-Aldrich, Dorset, United Kingdom) as required, and were grown at 30°C or 40°C for 3 days.
Mga2p-processing assay.
The full-length S. cerevisiae MGA2 ORF was cloned from yeast genomic DNA using PCR and inserted into a YCplac111 CEN LEU2-derived plasmid that adds an N-terminal HA epitope tag under the control of the TPI1 promoter. HA-Mga2p was visualized in equalized total protein extracts from log-phase yeast cultures using immunoblotting with monoclonal anti-HA antibodies as described above.
Microscopy.
Yeast containing mCherry-ΔNSmf1p was grown on metal-depleted medium as described previously (33). Similarly, yeast containing green fluorescent protein (GFP)-Gap1p was grown using 0.1% (wt/vol) proline as the sole nitrogen source (8) before imaging of log-phase cells. The CAN1 ORF was cloned from yeast genomic DNA using PCR and inserted into a YCplac33 CEN URA3-derived shuttle vector containing GFP (11). Yeast cells were grown to log phase in selective medium lacking arginine, which was added to the growth medium to a final concentration of 100 mg/liter 2 h before imaging. Fluorescent and differential interference contrast imaging of log-phase cells in water were performed using a Leica SP5 confocal microscope, and digital images were inverted and the brightness/contrast was altered for clarity using Photoshop CS4 software (Adobe, Mountain View, CA).
Metal depletion assays.
Yeast containing TAP2-ΔNSmf1p (the TAP2 tag is a variant of the standard Tandem Affinity Tag [33] in which the calmodulin binding domain has been replaced with a 6His region) were grown in metal-depleted medium as described previously (33). Log-phase cells were then grown for a further 16 h in either metal-depleted medium or metal-depleted medium to which 200 μM MnCl2 or 25 μM CdCl2 had been added. Total protein extracts were subjected to immunoblotting with peroxidase–anti-peroxidase antibodies (Sigma-Aldrich, Dorset, United Kingdom) and detection using chemiluminescence reagent (Millipore, Watford, United Kingdom).
Ubiquitination assays.
In vitro ubiquitination assays were performed as described previously (33) using recombinant GSTRsp5p and altered versions that had only a single functional WW domain (33). BUL ORFs were cloned into a pET30a vector (Merck, Nottingham, United Kingdom), and recombinant protein was produced in Escherichia coli strain BL21-CodonPlus (Stratagene, Stockport, United Kingdom) and purified using HIS-select resin (Sigma-Aldrich, Dorset, United Kingdom) according to the manufacturer's protocol.
RESULTS
S. cerevisiae has three BUL family genes.
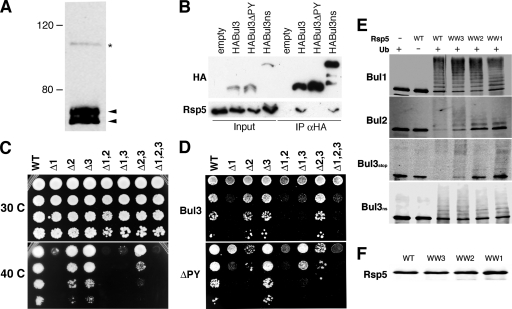
A BLAST search of the yeast genome identified two open reading frames encoding proteins with significant amino acid homology to Bul1p and Bul2p. Bsc5p (ynr069cp) and ynr068cp show homology to the N and C termini of Bul1p, respectively. Further examination found that these open reading frames are adjacent to each other in the genome and are separated by a stop codon that undergoes bypass translation at a reported level of ∼5% (23). The Bsc5p/ynr068cp protein (which we refer to here as Bul3p) shows significant identity and similarity to Bul1p and Bul2p (Table 1), including the presence of a putative PY motif (PPFY100) and a short arrestin-like motif (25). The Bul3p coding sequence was cloned from yeast genomic DNA, and the presence of the stop codon was confirmed through sequencing. Using an HA epitope-tagged Bul3p construct, we observed at least two distinct translation products of approximately 55 to 60 kDa, likely corresponding to modified forms of the Bsc5p portion of Bul3p. However, a larger protein approximately 100 kDa in size was also detected, albeit in much reduced abundance, corresponding to the bypass translation product (Fig. 1A). Both the large and small Bul3p proteins could be coimmunoprecipitated with Rsp5p from yeast cell extracts in a PY motif-dependent fashion (Fig. 1B).
Table 1.
Percent identity/similarity of the Bul proteinsa
| Protein | % Identity/similarity to: |
||
|---|---|---|---|
| Bul1 | Bul2 | Bul3 | |
| Bul1 | 51/66 | 31/50 | |
| Bul2 | 51/66 | 30/51 | |
| Bul3 | 31/50 | 30/51 | |
The table compares the global percent amino acid identity and similarity of Bul1p, Bul2p, and Bul3p (including the bypass stop codon).
Fig 1.
BUL genes are not essential for viability but are important for response to heat stress. (A) HA epitope-tagged Bul3p was expressed in a bul3Δ null mutant strain, and the protein was visualized by immunoblotting using anti-HA antibodies. The arrowheads indicate two major translation products of 55 to 60 kDa; the asterisk indicates a readthrough translation product of ∼100 kDa. (B) HABul3p was expressed in a bul3Δ strain, along with a Y100A (HABul3ΔPY) form, a variant that lacks the internal stop codon (HABul3ns), and an empty HA vector control. Extracts from log-phase cells (Input) were subjected to immunoprecipitation with anti-HA affinity resin (IP αHA) and subjected to immunoblotting with anti-HA and anti-Rsp5 antibodies. (C) Tenfold serial dilution of yeast strains with the BUL genes deleted alone (Δ1, Δ2, and Δ3), in pairs (Δ1,2; Δ1,3; and Δ2,3), or in combination (Δ1,2,3) were grown for 3 days at either 30°C or 42°C. WT, wild type. (D) Serial deletions as described for panel C with strains expressing HABul3 or HABul3ΔPY constructs. (E and F) In vitro ubiquitination assays using recombinant S-tagged Bul1p and Bul2p and large (Bul3ns) and small (Bul3stop) forms of Bul3p. The reactions were performed with methylated ubiquitin (Ub) using equal quantities, as determined using immunoblots with anti-Rsp5p antibodies (F), of wild-type Rsp5p and variants that had only a single functional WW domain remaining (WW3, WW2, and WW1) before being subjected to immunoblotting with anti-S-tag antibodies.
Bul proteins are not essential for cell viability.
Previous studies have shown that Bul1p and Bul2p bind to and regulate the activity of the ubiquitin ligase Rsp5p (35, 36). However, unlike Rsp5p, deletion of Bul1p and/or Bul2p has no effect on the viability of yeast cells (36). To determine if the survival of yeast cells in the absence of Bul1p and Bul2p was due to the activity of Bul3p, we made a BUL3 null mutation (see Materials and Methods) and examined the effect of this deletion in combination with loss of BUL1 and/or BUL2. Figure 1C shows that deletion of BUL3 has no effect on the growth of yeast cells at 30°C, either alone or in combination with the loss of BUL1 and/or BUL2. As reported previously (36), a bul1Δ mutant shows very restricted growth at 40°C while bul2Δ and bul3Δ mutants show only a slight temperature sensitivity, which is enhanced when both genes are absent (Fig. 1C). The temperature sensitivity of the bul2Δ bul3Δ double mutant could be complemented by overexpression of epitope-tagged Bul3p only when the PY motif was present (Fig. 1D). We also observed that overexpression of Bul3p was not able to complement the extreme temperature sensitivity phenotype of bul1Δ (Fig. 1D), and this was also seen in Bul3p constructs that lacked the internal stop codon (T. V. Novoselova and J. A. Sullivan, unpublished data). Interestingly, overexpression of Bul3p without a PY motif greatly enhanced the temperature sensitivity of bul2Δ (Fig. 1D), suggesting a possible regulatory interaction between Bul2p and Bul3p proteins independent of Rsp5p. Taken together, these observations demonstrate that while the Bul family proteins are not essential for cell viability, Bul3p has a role in combination with other members of the family in regulating the yeast cell's response to heat stress.
Bul PY motifs interact differently with Rsp5p.
Since a clear PY motif-dependent interaction of Bul3p with Rsp5p was observed in yeast, we decided to investigate the roles of the Rsp5p WW domains in this interaction using in vitro ubiquitination assays (Fig. 1E). As expected, recombinant Rsp5p was able to modify all of the Bul proteins examined, including both forms of Bul3p. Interestingly, in our assays, the small form of Bul3p (Fig. 1E, Bul3stop) was much more readily modified than the large form (Fig. 1E, Bul3ns), as evidenced by the complete loss of the unmodified protein and the appearance of only high-molecular-weight conjugates. We were also surprised to note that when we used single-WW-domain mutants of Rsp5p, the Bul protein PY motifs had different preferences for WW domains. While Bul1p interacts with equal efficiency with all WW domains, Bul2p has a preference for WW3 (reflected in reduced modification when WW1 is absent). In contrast, the small form of Bul3p preferentially interacts with WW2, while the large form interacts equally well with all WW domains (Fig. 1E). These results suggest that, in vitro at least, the Bul proteins interact differently with Rsp5p, which could lead to different functional outcomes.
The Bul protein family can be functionally antagonistic.
Since a temperature sensitivity phenotype was observed in a bul3Δ null mutant we decided to investigate the effects of other chemical agents previously linked to Rsp5p and Bul1p and/or Bul2p on yeast growth. Figure 2A and B show the effects on growth when cadmium, phleomycin, ethanol, and the amino acid analogs l-azetidine-2-carboxylic acid and canavanine were added to growth media.
Fig 2.
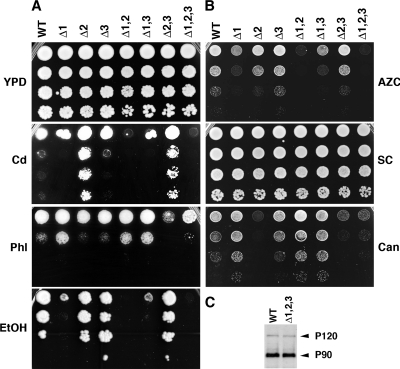
BUL gene deletions show complex growth phenotypes but are not required for ubiquitin-mediated processing of Mga2p. (A) Tenfold serial dilution of BUL gene deletion strains grown on YPD agar medium, YPD plus 25 μM CdCl2 (Cd) or 3 μM phleomycin (Phl), or YEP plus 2% (vol/vol) ethanol (EtOH). (B) Tenfold serial dilution of BUL gene deletion strains grown on YPD agar medium plus 1 mM AZC, synthetic complete medium with no arginine (SC), or SC minus Arg plus 14 μM canavanine (Can). (C) Immunoblot using anti-HA antibodies of HA-Mga2p processing from P120 to P90 in wild-type and BUL gene deletion (Δ1,2,3) strains.
Cadmium toxicity is mediated by the presence of the manganese transporter Smf1p at the yeast plasma membrane. In the presence of cadmium, Smf1p is ubiquitinated and endocytosed in a process requiring Rsp5p, preventing heavy metal entry into the cell and reducing toxicity (11, 22). Deletion of BUL1 makes yeast growth very sensitive to cadmium, while, surprisingly, loss of BUL2 makes yeast hyperresistant to cadmium (Fig. 2A, Cd) an observation similar to that of a yeast strain lacking SMF1 (Novoselova and Sullivan, unpublished). In contrast, loss of BUL3 has no effect on yeast growth in the presence of cadmium (Fig. 2A).
Bul1p has been shown previously to antagonize the Bre5p-Ubp3p protease complex in regulating response to damaged DNA (3). As reported previously (3) we observed that deletion of BUL1 increased the resistance of yeast cells to the DNA-damaging agent phleomycin (Fig. 2A, Phl). However, we also observed that loss of either BUL2 or BUL3 caused a slight increase in sensitivity to DNA damage while loss of both together made the yeast cells hypersensitive to phleomycin. Interestingly, when all three Bul proteins were absent, growth on phleomycin was not dissimilar to that observed with wild-type yeast, suggesting that increased sensitivity to phleomycin, caused by the loss of BUL2 and BUL3, has been compensated for by the lack of BUL1 (Fig. 2A, Phl).
Differing roles for the Bul proteins were again observed when null mutant strains were grown in the presence of ethanol (Fig. 2A, EtOH). Excess ethanol causes the activation of stress responses in yeast, and cell survival requires the Rsp5p-mediated degradation of misfolded proteins (12). Interestingly, while Bul1p appears to be required for this process, as concluded from the lack of growth of the bul1Δ strain in the presence of ethanol, loss of either BUL2 or BUL3 alone or in combination appears to have no effect on yeast viability (Fig. 2A). The lack of a role for Bul2p and Bul3p in ethanol-induced stress survival differs from what is observed with temperature-induced stress (Fig. 1C) and implies different roles for the Bul family depending upon the nature of the stress-inducing signal.
To investigate further the phenotypes of BUL family deletions, we examined the effects of AZC and canavanine on yeast growth. AZC and canavanine are amino acid analogs of proline and arginine, respectively. In both cases, the respective amino acid transporters for these analogs (Put4p and Can1p) are regulated by ubiquitin-mediated endocytosis involving Rsp5p (20). Deletion of BUL1 and, to a lesser extent, BUL2 made yeast cells more sensitive to the proline analog AZC (Fig. 2B, AZC). Deletion of BUL3 had no effect on the ability of yeast to grow in the presence of AZC, while deletion of BUL1 and BUL2 together prevented almost all growth. In contrast, only deletion of BUL2 made yeast cells sensitive to the arginine analog canavanine, with slight resistance to canavanine being observed when BUL1 was deleted (Fig. 2B, Can). Loss of BUL3 alone had little effect on the growth of yeast cells in the presence of canavanine; however, the bul1Δ bul2Δ bul3Δ strain was much more sensitive to canavanine than the bul1Δ bul2Δ strain, suggesting some potential role for BUL3 in regulating the activity of Can1p.
The Bul family is not involved in all Rsp5p-dependent processes.
To investigate the extent of the Bul family involvement in cellular processes controlled by Rsp5p, we examined the effects of deleting the Bul proteins on the processing of the Mga2p transcription factor. Mga2p and Sgt23p are PY motif-containing transcription factors that are translated as proproteins with an integral membrane domain causing their retention in the yeast endoplasmic reticulum (ER). Following ubiquitination by Rsp5p, these proteins are partially processed by the 26S proteasome, releasing a soluble transcription factor (13, 28). To investigate the roles of the Bul proteins in this ubiquitin-mediated proteolytic-processing event, we examined the processing of an HA-epitope-tagged Mga2p construct from its P120 unprocessed form to the smaller P90 form. As can been seen in Fig. 2C, in wild-type cells, approximately 60% of the HA-Mga2p protein is found in the P90 processed form, and deletion of all BUL genes had no significant effect on the extent to which P120 is processed to P90. Similar results were obtained with single and double deletions of BUL genes (Novoselova and Sullivan, unpublished), indicating that not all of the Rsp5p-dependent processes in the yeast cell require a member of the Bul family of proteins.
Bul proteins differentially affect sorting of plasma membrane proteins.
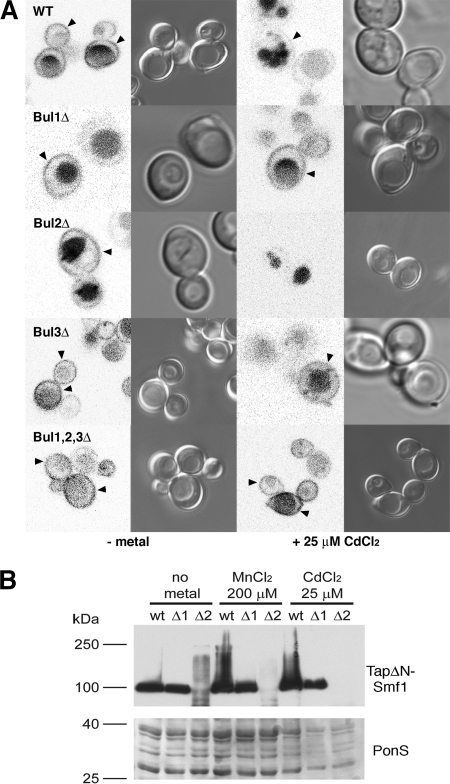
Since loss of Bul1p made cells hypersensitive to growth on cadmium while loss of Bul2p made cells hyperresistant, we decided to investigate the localization of the plasma membrane transporter Smf1p in BUL family mutants. Two modes of Rsp5p ubiquitin-mediated sorting of Smf1p have been reported, a stress-triggered endocytosis event that requires Rsp5p and the arrestin-like proteins Ecm21p (Art2p) and Csr2p (Art8p) (26) and a metal-dependent ubiquitin-mediated sorting event that requires Rsp5p and the adaptor proteins Bsd2p and Tre1/2p (32, 33). To investigate this system further, we examined the localization of mCherry-tagged ΔNSmf1p. ΔNSmf1p has a deletion of the N terminus of Smf1p that prevents stress-induced endocytosis but still allows ubiquitin-mediated metal-dependent sorting (33). As reported previously (33), growth of yeast cells in metal-depleted medium caused mCherry-ΔNSmf1p to accumulate at the plasma membrane (Fig. 3A), although significant quantities of mCherry-ΔNSmf1p are trafficked to the vacuole, reflected in the intense vacuolar fluorescent signal (caused by the relative resistance of mCherry to vacuolar proteases). Deletion of the BUL genes alone or in combination had no effect on the plasma membrane localization of mCherry-ΔNSmf1p in metal-depleted medium, although a reduced vacuolar signal was observed in strains that lack BUL3 (Fig. 3A). Following the addition of cadmium, no mCherry-ΔNSmf1p was observed at the plasma membrane in a bul2Δ strain, in contrast to a wild-type, bul1Δ, bul3Δ, or bul1Δ bul2Δ bul3Δ strain, where plasma membrane localization of mCherry was still identifiable (Fig. 3A). This enhanced sorting of ΔNSmf1p in a bul2Δ strain is also reflected in immunoblots of extracts from yeast strains expressing a TAP-tagged ΔNSmf1p (Fig. 3B). When grown on metal-depleted medium, a single band corresponding to TAP-ΔNSmf1p was observed in a wild-type and a bul1Δ yeast; in contrast, only highly modified TAP-tagged ΔNSmf1p, likely to be ubiquitinated forms, was observed in a bul2Δ strain (Fig. 2B). Addition of a large excess of either manganese or cadmium results in the disappearance of TAP-ΔNSmf1p in the bul2Δ strain and the accumulation of modified forms of the protein in a wild-type strain but no change in the bul1Δ strain.
Fig 3.
Sorting of mCherryΔNSmf1p in metal-depleted medium following the addition of CdCl2. (A) mCherry fluorescence and differential interference contrast microscopy images of wild-type and single BUL gene deletion strains expressing mCherryΔNSmf1p following growth on metal-depleted media (−metal) and 2 h after the addition of 25 μM CdCl2. The arrowheads indicate plasma membrane localization. (B) (Top) Immunoblots against Tap-tagged ΔNSmf1p in total protein extracts from wild-type and bul1Δ (Δ1) and bul2Δ (Δ2) strains. (Bottom) PonceauS staining was used as a loading control.
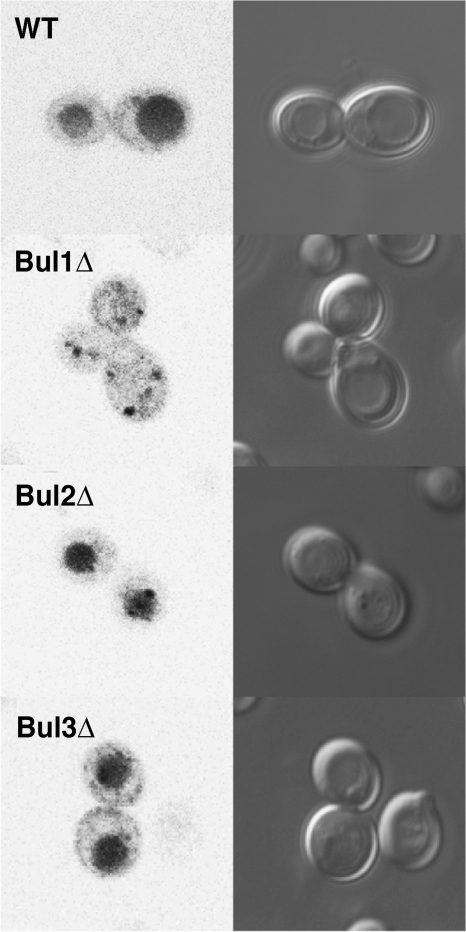
To further examine the roles of the Bul proteins in ubiquitin-mediated sorting, we also investigated the localization of the general amino acid permease Gap1p in nitrogen-poor medium. The ubiquitin-mediated endocytosis of Gap1p in response to nitrogen has been shown previously to require Rsp5p, Bul1p, and Bul2p (10). In contrast to previously published results, we observed very little plasma membrane localization of GFP-Gap1p in wild-type yeast cells, with the majority of the GFP being found in the yeast vacuole and only a small amount of GFP signal seen at the cell periphery (Fig. 4). This is likely due to our use of the BY4742 yeast strain rather than S288C, which has altered Gap1p expression and has been used previously to visualize plasma membrane localization of Gap1p (10). In our hands, only deletion of BUL1 caused a significant change in the localization of GFP-Gap1p, with GFP being restricted to intracellular puncta and likely to yeast endosomes/Golgi network (Fig. 4). Surprisingly, deletion of BUL2 or BUL3 showed very little difference in GFP-Gap1p localization from that observed in wild-type yeast.
Fig 4.
Sorting of GFPGap1p in nitrogen-poor medium. Shown are GFP fluorescence and differential interference contrast microscopy images of wild-type and single BUL gene deletion strains expressing GFPGap1p following growth in medium with proline as the sole nitrogen source.
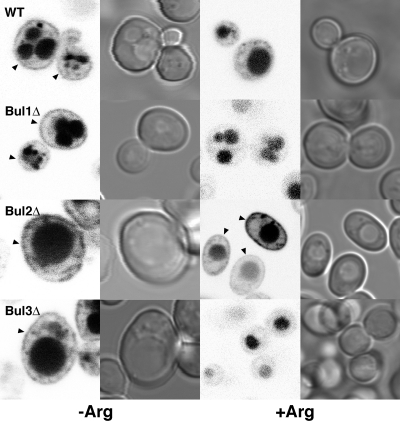
The ubiquitin-mediated endocytosis of the arginine transporter Can1p in the presence of excess arginine has been shown previously to involve Rsp5p and the arrestin-like protein Art1p (20). Since we had observed differential sensitivity of BUL gene deletions to the arginine analog canvanine (Fig. 2B), we also investigated the localization of GFP-Can1p. When grown in the absence of arginine, a clear GFP signal was observed at the cell periphery in all strains examined (Fig. 5). However, following the addition of arginine, only in a BUL2 deletion strain were significant quantities of GFP-Can1p observed at the cell periphery (Fig. 5). In all strains investigated, including the bul2Δ strain, a significant GFP signal was observed in the yeast vacuole, suggesting that sorting of Can1p via the MVB pathway was unimpaired.
Fig 5.
Sorting of GFPCan1p following growth in excess arginine. Shown are GFP fluorescence and differential interference contrast microscopy images of wild-type and single BUL gene deletion strains expressing GFPCan1p. The images were obtained before (−Arg) and 2 h after (+Arg) the addition of 100 mg/liter arginine to log-phase cells grown in arginine-free synthetic complete medium. The arrowheads indicate plasma membrane localization.
These results show that different members of the Bul family regulate the sorting of Smf1p, Gap1p, and Can1p, but not the ubiquitin-mediated processing of Mga2p. Primarily, this regulation appears to involve Bul1p and Bul2p for Smf1p and Gap1p, although there is some role for Bul3p in the sorting of Can1p.
DISCUSSION
Understanding how E3 ligases like Rsp5p are regulated is critical to understanding the process of ubiquitination (14, 18, 29). Here, we report the identification of a new Rsp5p regulatory protein, Bul3p, which, along with the other members of the Bul family, regulates some of the cellular processes involving Rsp5p. We also report the surprising observation that members of the Bul family can show antagonistic activity toward the same Rsp5p substrate. This is best seen in the ubiquitin-mediated sorting of the manganese transporter Smf1p, where Bul1p and Bul2p have opposite roles in controlling ubiquitin-mediated sorting. Our results suggest that while Bul1p is required for Smf1 endocytosis, through recruitment and subsequent ubiquitination by Rsp5p, Bul2p inhibits this process. Similarly, Bul1p and Bul2p appear to antagonize each other with respect to the ubiquitin-mediated sorting of the arginine transporter Can1p. However, in this case, Bul2p appears to enhance Can1p sorting while Bul1p has the opposite function. Further complexity in the roles of the Bul proteins is seen in the sorting of the permease Gap1p, which requires only Bul1p with no apparent role for either Bul2p or Bul3p, contradicting previous studies (10). It is possible that these differences reflect the use of a different yeast strain or, more likely, are based on the incorrect assumption of Bul1p and Bul2p functional redundancy. However, it is clear that the role of the Bul family in ubiquitin-mediated sorting of plasma membrane proteins is not universal. It is not clear at this stage if there is a role for Bul proteins in ubiquitin-mediated endocytosis and/or ubiquitin-mediated diversion of Smf1p, Can1p, and Gap1p from the Golgi apparatus/endosome. However, given our observations and those of others (31) with Smf1p and Gap1p, roles for Bul proteins in both these processes seem possible.
Although we have yet to identify a unique role for Bul3p in ubiquitin-mediated sorting of plasma membrane proteins, Bul3p does appear to be particularly important for cell survival following DNA damage after phleomycin treatment. Previous studies have shown that Bul1p functions antagonistically to the Bre5p-Ubp3p ubiquitin protease complex to regulate nonhomologous-end-joining (NHEJ) DNA repair mechanisms (3). Our results suggest that like Bre5p/Ubp3p, Bul2p or Bul3p can antagonize Bul1p following DNA damage. A role for Bre5p/Ubp3p in ER-to-Golgi apparatus trafficking has been demonstrated (5), and it has been shown that Bre5p/Ubp3p modulates the ubiquitin status of TFIID (1) and RNAPII (17). It is conceivable that the Bul proteins regulate Rsp5p activity on a particular target protein and that it is the ubiquitination status of this protein that is subsequently modulated by Bre5p/Ubp3p to ultimately control NHEJ pathways.
The presence of an internal stop codon in BUL3 that undergoes readthrough translation is an interesting conundrum. Numerous BUL-like genes have been identified in other fungal species, but we have been unable to find a similarly positioned stop codon. The low sequence identity between Bul proteins in S. cerevisiae and between Bul proteins in other species makes it difficult to conclusively identify Bul3p through homology searches alone. Indeed, it seems extremely likely that some Bul3p homologs have been misannotated as either Bul1p or Bul2p. However, a clear Bul3p homolog can be identified in Zygosaccharomyces rouxii, a product of the ORF ZYRO0G14058g, which shows higher identity/similarity (36/55%) to S. cerevisiae Bul3p than Bul3p shows to either S. cerevisiae Bul1p or Bul2p, and in this case, no internal stop codon is present. The reason for the internal stop codon in S. cerevisiae Bul3p remains unclear. It may represent a potential method of regulation and/or indicate that the two Bul3 proteins may have subtly different activities on some substrate(s) yet to be identified. Indeed, our experiments suggest that the two forms of Bul3p can interact with Rsp5p in different ways in vitro, potentially leading to different functional outcomes. At present, we have been unable to find a phenotypic difference between strains containing BUL3 with or without a stop codon. However, given the potential number of Rsp5p substrates in the yeast cell, the ubiquitination of any one of which may involve Bul3p, this is not too surprising.
One obvious question is, how do the Bul family proteins control Rsp5p activity? Previous studies with Gap1p have suggested that Bul1p and Bul2p function as E4 enzymes to extend polyubiquitin chains (10). However, Rsp5p alone is perfectly capable of attaching long polyubiquitin chains to its targets (33), and we observed no effect of Bul1p on the ability of Rsp5p to form polyubiquitin chains in vitro (Novoselova and Sullivan, unpublished). More recently, it has been suggested that Bul1p and Bul2p are distantly related arrestin-like adaptor proteins that recruit Rsp5p to substrates (25). However, our observation that Bul1p and Bul2p antagonize ubiquitin-mediated sorting of some substrates appears not to agree with a simple role for the Bul family as adaptor proteins. An alternate hypothesis, consistent with our observations, is that the Bul proteins function to regulate the adaptor-Rsp5p interaction. Given the large number of substrates and adaptors of Rsp5p, efficient association and disassociation with Rsp5p are essential for the ligase to function correctly. In an adaptor-regulator model, the Bul proteins would function as gatekeepers of the ligase-adaptor interaction, ensuring both efficient binding and removal of adaptors from the ligase. In this model, Bul1p would allow the efficient recruitment of adaptors, such as Art2p and Art8p, to facilitate ubiquitin-mediated endocytosis of Smf1p, while Bul2p would displace Art2/8p from Rsp5p to allow efficient “recycling” of the ligase for subsequent interactions. In this case, the Bul proteins could partially compensate for the absence of a primary adaptor for a particular substrate by enhancing the interaction between a substrate and a lower-affinity adaptor, explaining the apparent functional redundancy of Bul1p and Bul2p with the Art proteins observed previously (25). One simple mechanistic model to explain this Rsp5p gating activity suggested by our experiments is that by binding to Rsp5p, the Bul proteins may alter access to specific WW domains. Blocking access to a particular WW domain could prevent a productive interaction or alter the interaction of an Rsp5p/adaptor to a more effective conformation. Previous work (33) has shown that complex interactions can take place between adaptors and Rsp5p and that correct positioning of an adaptor on Rsp5p is often essential for activity. For example, the adaptor protein Bsd2p strongly interacts with the WW3 domain of Rsp5p, and this interaction is essential for the metal-dependent ubiquitination and sorting of Smf1p (33). In our experiments, Bul2p also binds preferentially to WW3, which may explain why in its absence Smf1p sorting appears to be more efficient.
It is interesting that no Bul homologs can be identified in higher eukaryotes, even though Nedd4 ligases are present. Of course, the presence of structural rather than sequence homologs cannot be ruled out. However, it is striking that higher eukaryotes tend to have multiple Nedd4 ligases (29), and it is possible that S. cerevisiae requires an extra level of regulation on Rsp5p provided by the Bul family that is not needed where multiple, more specialized Nedd4 ligases are found.
Understanding how Nedd4 ligases recognize their substrates and the involvement of adaptor protein substrate recognition is an interesting and complex problem. The Bul proteins may represent the answer to a fungus-specific question of how to control the activity of a single ubiquitin ligase that needs to be differently and efficiently regulated in multiple cellular locations.
ACKNOWLEDGMENTS
We thank Gina Devasahayam and Thomas Sturgill for the gift of the GFPGap1 plasmid, Elina Nikko for the BUL1 BUL2 double-mutant knockout strain, and Hugh Pelham and Richard Pickersgill for comments on the manuscript.
This work was supported by a research grant from the Biotechnology and Biological Science Research Council.
Footnotes
Published ahead of print 3 February 2012
REFERENCES
- 1. Auty R, et al. 2004. Purification of active TFIID from Saccharomyces cerevisiae. Extensive promoter contacts and co-activator function. J. Biol. Chem. 279:49973–49981 [DOI] [PubMed] [Google Scholar]
- 2. Bernassola F, Karin M, Ciechanover A, Melino G. 2008. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell 14:10–21 [DOI] [PubMed] [Google Scholar]
- 3. Bilsland E, Hult M, Bell S, Sunnerhagen P, Downs J. 2007. The Bre5/Ubp3 ubiquitin protease complex from budding yeast contributes to the cellular response to DNA damage. DNA Repair 6:1471–1484 [DOI] [PubMed] [Google Scholar]
- 4. Clague M, Urbé S. 2006. Endocytosis: the DUB version. Trends Cell Biol. 16:551–559 [DOI] [PubMed] [Google Scholar]
- 5. Cohen M, Stutz F, Belgareh N, Haguenauer-Tsapis R, Dargemont C. 2003. Ubp3 requires a cofactor, Bre5, to specifically de-ubiquitinate the COPII protein, Sec23. Nat. Cell Biol. 5:661–667 [DOI] [PubMed] [Google Scholar]
- 6. David Y, Ziv T, Admon A, Navon A. 2010. The E2 ubiquitin conjugating enzymes direct polyubiquitination to preferred lysines. J. Biol. Chem. 285:8595–8604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deshaies R, Joazeiro C. 2009. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78:399–434 [DOI] [PubMed] [Google Scholar]
- 8. Devasahayam G, Burke D, Sturgill T. 2007. Golgi manganese transport is required for rapamycin signaling in Saccharomyces cerevisiae. Genetics 177:231–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dunn R, Hicke L. 2001. Multiple roles for Rsp5p-dependent ubiquitination at the internalization step of endocytosis. J. Biol. Chem. 276:25974–25981 [DOI] [PubMed] [Google Scholar]
- 10. Helliwell S, Losko S, Kaiser C. 2001. Components of a ubiquitin ligase complex specify polyubiquitination and intracellular trafficking of the general amino acid permease. J. Cell Biol. 153:649–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hettema E, Valdez-Taubas J, Pelham H. 2004. Bsd2 binds the ubiquitin ligase Rsp5 and mediates the ubiquitination of transmembrane proteins. EMBO J. 23:1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hiraishi H, Okada M, Ohtsu I, Takagi H. 2009. A functional analysis of the yeast ubiquitin ligase Rsp5: the involvement of the ubiquitin-conjugating enzyme Ubc4 and poly-ubiquitination in ethanol-induced down-regulation of targeted proteins. Biosci. Biotechnol. Biochem. 73:2268–2273 [DOI] [PubMed] [Google Scholar]
- 13. Hoppe T. 2005. Multiubiquitylation by E4 enzymes: ‘one size’ doesn't fit all. Trends Biochem. Sci. 30:183–187 [DOI] [PubMed] [Google Scholar]
- 14. Hoppe T, et al. 2000. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell 102:577–586 [DOI] [PubMed] [Google Scholar]
- 15. Kanelis V, Bruce M, Skrynnikov N, Rotin D, Forman-Kay J. 2006. Structural determinants for high-affinity binding in a Nedd4 WW3* domain-Comm PY motif complex. Structure 14:543–553 [DOI] [PubMed] [Google Scholar]
- 16. Kee Y, Huibregtse J. 2007. Regulation of catalytic activities of HECT ubiquitin ligases. Biochem. Biophys. Res. Commun. 354:329–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kvint K, et al. 2008. Reversal of RNA polymerase II ubiquitylation by the ubiquitin protease Ubp3. Mol. Cell 30:498–506 [DOI] [PubMed] [Google Scholar]
- 18. Léon S, Haguenauer-Tsapis R. 2009. Ubiquitin ligase adaptors: regulators of ubiquitylation and endocytosis of plasma membrane proteins. Exp. Cell Res. 315:1574–1583 [DOI] [PubMed] [Google Scholar]
- 19. Léon S, Erpapazoglou Z, Haguenauer-Tsapis R. 2008. Ear1p and Ssh4p are new adaptors of the ubiquitin ligase Rsp5p for cargo ubiquitylation and sorting at multivesicular bodies. Mol. Biol. Cell 19:2379–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lin CH, Macgurn JA, Chu T, Stefan CJ, Emr SD. 2008. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell 135:714–725 [DOI] [PubMed] [Google Scholar]
- 21. Liu J, Sitaram A, Burd C. 2007. Regulation of copper-dependent endocytosis and vacuolar degradation of the yeast copper transporter, Ctr1p, by the Rsp5 ubiquitin ligase. Traffic 8:1375–1384 [DOI] [PubMed] [Google Scholar]
- 22. Liu X, Culotta V. 1999. Mutational analysis of Saccharomyces cerevisiae Smf1p, a member of the Nramp family of metal transporters. J. Mol. Biol. 289:885–891 [DOI] [PubMed] [Google Scholar]
- 23. Namy O, et al. 2003. Identification of stop codon readthrough genes in Saccharomyces cerevisiae. Nucleic Acids Res. 31:2289–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Navon A, Ciechanover A. 2009. The 26 S proteasome: from basic mechanisms to drug targeting. J. Biol. Chem. 284:33713–33718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nikko E, Pelham H. 2009. Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic 10:1856–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nikko E, Sullivan J, Pelham H. 2008. Arrestin-like proteins mediate ubiquitination and endocytosis of the yeast metal transporter Smf1. EMBO Rep. 9:1216–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pickart C. 2004. Back to the future with ubiquitin. Cell 116:181–190 [DOI] [PubMed] [Google Scholar]
- 28. Rape M, et al. 2001. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell 107:667–677 [DOI] [PubMed] [Google Scholar]
- 29. Shearwin-Whyatt L, Dalton H, Foot N, Kumar S. 2006. Regulation of functional diversity within the Nedd4 family by accessory and adaptor proteins. BioEssays 28:617–628 [DOI] [PubMed] [Google Scholar]
- 30. Slagsvold T, Pattni K, Malerød L, Stenmark H. 2006. Endosomal and non-endosomal functions of ESCRT proteins. Trends Cell Biol. 16:317–326 [DOI] [PubMed] [Google Scholar]
- 31. Soetens O, De Craene JO, Andre B. 2001. Ubiquitin is required for sorting to the vacuole of the yeast general amino acid permease, Gap1. J. Biol. Chem. 276:43949–43957 [DOI] [PubMed] [Google Scholar]
- 32. Stimpson H, Lewis M, Pelham H. 2006. Transferrin receptor-like proteins control the degradation of a yeast metal transporter. EMBO J. 25:662–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sullivan J, Lewis M, Nikko E, Pelham H. 2007. Multiple interactions drive adaptor-mediated recruitment of the ubiquitin ligase rsp5 to membrane proteins in vivo and in vitro. Mol. Biol. Cell 18:2429–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Welchman R, Gordon C, Mayer R. 2005. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell. Biol. 6:599–609 [DOI] [PubMed] [Google Scholar]
- 35. Yashiroda H, Oguchi T, Yasuda Y, Toh-EA , Kikuchi Y. 1996. Bul1, a new protein that binds to the Rsp5 ubiquitin ligase in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:3255–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yashiroda H, Kaida D, Toh-e A, Kikuchi Y. 1998. The PY motif of Bul1 protein is essential for growth of Saccharomyces cerevisiae under various stress conditions. Gene 225:39–46 [DOI] [PubMed] [Google Scholar]