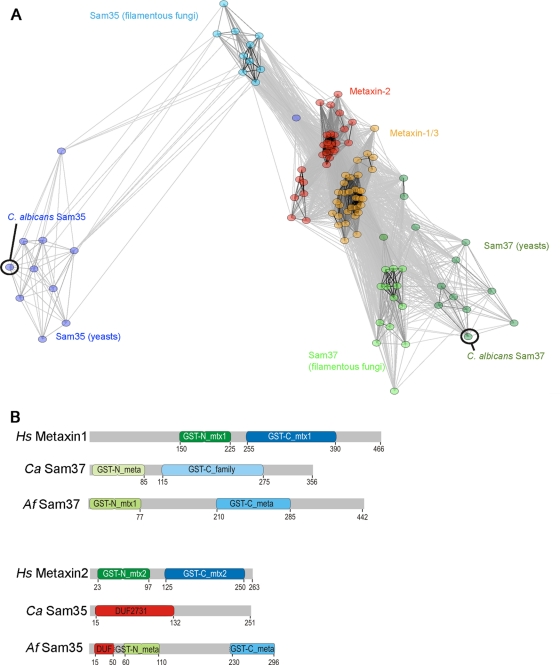
Fig 6.
Bioinformatic analysis of the fungal Sam37 and Sam35 proteins and the metaxins. (A) Sequence similarity between Sam37, Sam35, and metaxins, visualized using CLANS (18). Sequences are clustered based on the pairwise P value between two sequences in an all-against-all BLAST search. Circles represent individual sequences, while lines are drawn between any two sequences that have P values of <10−6. (B) Domain structure of the metaxins and the fungal Sam35 and Sam37 proteins from C. albicans and Aspergillus fumigatus. The indicated domains are from the conserved domain database, in which one of the major groupings is the glutathione S-transferase (GST) family, to which the metaxins and the SAMs belong. The presence of a conserved N-terminal domain with a thioredoxin fold casts a protein in the GST-N_family, while the presence of a conserved C-terminal alpha helical domain casts it in a GST-C_family. GST-N_meta(xin) and GST-C_meta(xin) are conserved subgroups of the GST-N and GST-C families, respectively, which include many of the metaxin/SAM subunits. Within the “meta” families, the proteins most conserved with metaxin 1 from animals are designated “mtx1” (e.g., GST_N_mtx1), and those most conserved with metaxin 2 are called “mtx2.” CaSam35 does not conform to the GST family but, like many other Sam35 homologs, it has conserved domain features classified as DUF2731 (DUF is domain of unknown function). DUF2731 must share some sequence-based features with the GST-N_family, given that AfSam35 partly conforms to the DUF2731 group and partly to the GST-N_meta(xin) group of sequences. We attempted homology modeling of Sam35, Sam37, and the metaxins; however, no reliable models could be made based on template proteins with a GST-like fold that we used.